Difference between revisions of "SpoIIIAH"
(→Labs working on this gene/protein) |
|||
| Line 37: | Line 37: | ||
=== Basic information === | === Basic information === | ||
| − | * ''' | + | * '''Locus tag:''' |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
Revision as of 00:50, 3 June 2009
- Description: required for SigG activation
| Gene name | spoIIIAH |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | activation of SigG |
| MW, pI | 23 kDa, 4.627 |
| Gene length, protein length | 654 bp, 218 aa |
| Immediate neighbours | accB, spoIIIAG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 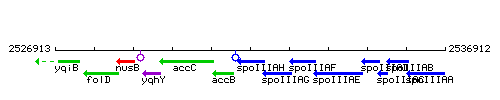
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: membrane protein PubMed
Database entries
- Structure:
- Swiss prot entry: P49785
- KEGG entry: BSU24360
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Additional information: the internal promoter is essential for sporulation, it is twice as active as the promoter in front of spoIIIAA, suggesting that SpoIIIAG and SpoIIIAH are required in larger amounts as compared to the other products of the operon PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed