Difference between revisions of "AprE"
| Line 1: | Line 1: | ||
| − | + | * '''Description:''' major extracellular alkaline protease <br/><br/> | |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 117: | Line 113: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=aprE_1104423_1105568_-1 aprE] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=aprE_1104423_1105568_-1 aprE] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|2447063}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|2447063}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 154: | Line 150: | ||
<pubmed>20735481 </pubmed> | <pubmed>20735481 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed> 19710937 3082852,2447063,2447062,19251843,18414485,6322190,12884008,15126467,1898931,1906467,2504584,18957862 11101663 12055299, 3131303 1906467 15598897 11325965 14688235 21099137 22745669 </pubmed> | + | <pubmed> 19710937 3082852,2447063,2447062,19251843,18414485,6322190,12884008,15126467,1898931,1906467,2504584,18957862 11101663 12055299, 3131303 1906467 15598897 11325965 14688235 21099137 22745669 23563549 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:26, 15 April 2013
- Description: major extracellular alkaline protease
| Gene name | aprE |
| Synonyms | sprE |
| Essential | no |
| Product | extracellular alkaline serine protease (subtilisin E)) |
| Function | protein degradation |
| Gene expression levels in SubtiExpress: aprE | |
| Interactions involving this protein in SubtInteract: AprE | |
| MW, pI | 39 kDa, 9.342 |
| Gene length, protein length | 1143 bp, 381 aa |
| Immediate neighbours | yhfN, yhfO |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 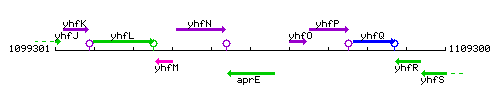
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed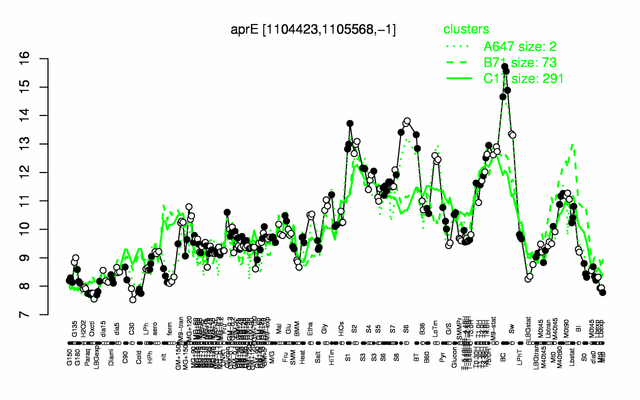
| |
Contents
Categories containing this gene/protein
utilization of nitrogen sources other than amino acids
This gene is a member of the following regulons
AbrB regulon, ScoC regulon, SinR regulon
The gene
Basic information
- Locus tag: BSU10300
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of proteins with broad specificity for peptide bonds, and a preference for a large uncharged residue in P1 (according to Swiss-Prot)
- Protein family: peptidase S8 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: secreted (according to Swiss-Prot), extracellular (signal peptide) PubMed
Database entries
- Structure: 1SBC
- UniProt: P04189
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: aprE (according to DBTBS)
- Regulatory mechanism:
- Additional information: the mRNA is extremely stable (more than 25 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
- Additional information: The pAPNC213 vector published by Morimoto et al. (2002) is an insertion plasmid for IPTG inducible constructs that can be integrated into the chromosomal aprE locus by replacing the aprE open reading frame via double homologous recombination. PubMed
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications