Difference between revisions of "Nin"
| Line 31: | Line 31: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nin_371729_372127_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:nin_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=nin_371729_372127_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:nin_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU03420]] |
|- | |- | ||
|} | |} | ||
Revision as of 12:31, 16 May 2013
- Description: inhibitor of the DNA degrading activity of NucA
| Gene name | nin |
| Synonyms | comJ |
| Essential | no |
| Product | inhibitor of the DNA degrading activity of NucA |
| Function | genetic transformation, DNA uptake |
| Gene expression levels in SubtiExpress: nin | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 14 kDa, 4.922 |
| Gene length, protein length | 396 bp, 132 aa |
| Immediate neighbours | yckE, nucA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed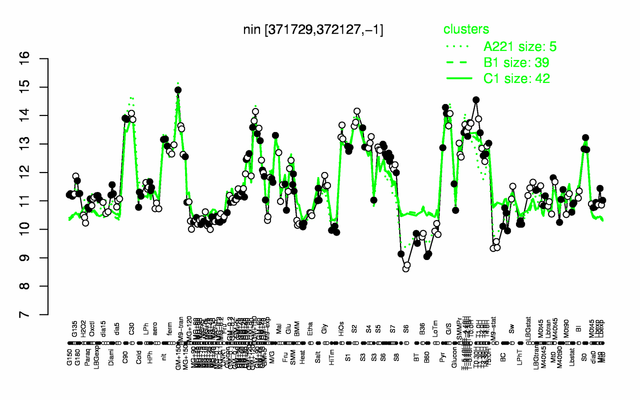
| |
Contents
Categories containing this gene/protein
genetic competence, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU03420
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- phosphorylated on Arg-96 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P12669
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Hanne Jarmer, Randy Berka, Steen Knudsen, Hans H Saxild
Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions.
FEMS Microbiol Lett: 2002, 206(2);197-200
[PubMed:11814663]
[WorldCat.org]
[DOI]
(P p)
R Provvedi, I Chen, D Dubnau
NucA is required for DNA cleavage during transformation of Bacillus subtilis.
Mol Microbiol: 2001, 40(3);634-44
[PubMed:11359569]
[WorldCat.org]
[DOI]
(P p)
D van Sinderen, R Kiewiet, G Venema
Differential expression of two closely related deoxyribonuclease genes, nucA and nucB, in Bacillus subtilis.
Mol Microbiol: 1995, 15(2);213-23
[PubMed:7746143]
[WorldCat.org]
[DOI]
(P p)