Difference between revisions of "SpoIVCB"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 110: | Line 110: | ||
* '''Operon:''' ''[[sigK]]'' (composed of ''[[spoIVCB]]-[[spoIIIC]]'') {{PubMed|2536191}} | * '''Operon:''' ''[[sigK]]'' (composed of ''[[spoIVCB]]-[[spoIIIC]]'') {{PubMed|2536191}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=spoIVCB_2652993_2653463_1 spoIVCB] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigE]] {{PubMed|15699190,2841290}}, [[SigK]] {{PubMed|2841290}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 11:47, 16 April 2012
- Description: sporulation-specific sigma factor (SigK) (N-terminal half), other part: spoIIIC
| Gene name | spoIVCB |
| Synonyms | cisB, sigK |
| Essential | no |
| Product | RNA polymerase sporulation-specific sigma factor (sigma-K) (N-terminal half) |
| Function | late mother cell-specific gene expression |
| Interactions involving this protein in SubtInteract: SigK | |
| MW, pI | 17 kDa, 8.208 |
| Gene length, protein length | 468 bp, 156 aa |
| Immediate neighbours | nucB, spoIVCA |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 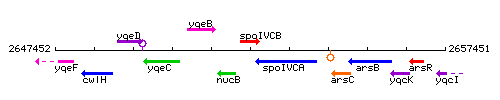
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, sporulation proteins, membrane proteins
This gene is a member of the following regulons
GerE regulon, SigE regulon, SigK regulon, SpoIIID regulon
The SigK regulon
The gene
Basic information
- Locus tag: BSU25760
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P12254
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Arthur Aronson, Purdue University, West Lafayette, USA homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
The SigK regulon
Takeko Kodama, Takeshi Matsubayashi, Tadayoshi Yanagihara, Hiroyuki Komoto, Katsutoshi Ara, Katsuya Ozaki, Ritsuko Kuwana, Daisuke Imamura, Hiromu Takamatsu, Kazuhito Watabe, Junichi Sekiguchi
A novel small protein of Bacillus subtilis involved in spore germination and spore coat assembly.
Biosci Biotechnol Biochem: 2011, 75(6);1119-28
[PubMed:21670523]
[WorldCat.org]
[DOI]
(I p)
Jessica M Silvaggi, John B Perkins, Richard Losick
Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis.
J Bacteriol: 2006, 188(2);532-41
[PubMed:16385044]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
Original Publications