Difference between revisions of "AddB"
(→References) |
|||
| Line 54: | Line 54: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| + | ** the enzyme is functional as a heterodimer of the [[AddA]] and [[AddB]] subunits, that it is a rapid and processive DNA helicase, and that it catalyses DNA unwinding using one single-stranded DNA motor of 3'→5' polarity located in the [[AddA]] subunit {{PubMed|21071401}} | ||
| + | ** the [[AddB]] subunit contains a second putative ATP-binding pocket, but this does not contribute to the observed helicase activity and may instead be involved in the recognition of recombination hotspot sequences {{PubMed|21071401}} | ||
* '''Protein family:''' uvrD-like helicase C-terminal domain (according to Swiss-Prot) | * '''Protein family:''' uvrD-like helicase C-terminal domain (according to Swiss-Prot) | ||
| Line 122: | Line 124: | ||
<pubmed>, 20116346 </pubmed> | <pubmed>, 20116346 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>,8387145,15610857,7746142, 19129187 1646786 10756102 9781875 17570399 20350930 </pubmed> | + | <pubmed>,8387145,15610857,7746142, 19129187 1646786 10756102 9781875 17570399 20350930 21071401 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:16, 13 November 2010
- Description: ATP-dependent deoxyribonuclease (subunit B)
| Gene name | addB |
| Synonyms | |
| Essential | no |
| Product | ATP-dependent deoxyribonuclease (subunit B)) |
| Function | DNA repair and recombination |
| MW, pI | 134 kDa, 5.39 |
| Gene length, protein length | 3498 bp, 1166 aa |
| Immediate neighbours | yhjR, addA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 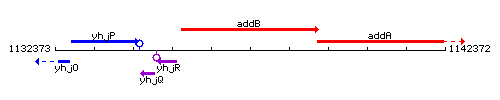
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU10620
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- the enzyme is functional as a heterodimer of the AddA and AddB subunits, that it is a rapid and processive DNA helicase, and that it catalyses DNA unwinding using one single-stranded DNA motor of 3'→5' polarity located in the AddA subunit PubMed
- the AddB subunit contains a second putative ATP-binding pocket, but this does not contribute to the observed helicase activity and may instead be involved in the recognition of recombination hotspot sequences PubMed
- Protein family: uvrD-like helicase C-terminal domain (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 1Y26 (the riboswitch in complex with adenine)
- UniProt: P23477
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP1106 (addAB, spc), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications