Difference between revisions of "ClpC"
| Line 149: | Line 149: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' ''[[clpC]]::tet'' available from the [http://subtiwiki.uni-goettingen.de/wiki/index.php/Leendert_Hamoen Hamoen] Lab | + | * '''Mutant:''' |
| + | ** ''[[clpC]]::tet'' available from the [http://subtiwiki.uni-goettingen.de/wiki/index.php/Leendert_Hamoen Hamoen] Lab | ||
| + | ** BP98 (''clpP''::''spc''), available in [[Fabian Commichau]]'s lab {{PubMed|25610436}} | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 162: | Line 164: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| − | [[Leendert Hamoen]], Newcastle University, UK [http://www.ncl.ac.uk/camb/staff/profile/l.hamoen homepage] | + | * [[Leendert Hamoen]], Newcastle University, UK [http://www.ncl.ac.uk/camb/staff/profile/l.hamoen homepage] |
| − | + | * [[Kürsad Turgay]], Freie Universität Berlin, Germany [http://www.biologie.fu-berlin.de/en/microbio/mibi-turg/index.html homepage] | |
| − | [[Kürsad Turgay]], Freie Universität Berlin, Germany [http://www.biologie.fu-berlin.de/en/microbio/mibi-turg/index.html homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 172: | Line 173: | ||
<pubmed> 17302811 23375660 23479438,19609260,19781636</pubmed> | <pubmed> 17302811 23375660 23479438,19609260,19781636</pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>9987115,8016067,9000055,12923101,10447896,9141693,2113920,16497325,19226326,8793870,10809708,14679237,17560370,11684022,8195092,11722737,11914365,12028382,18689476,19361434,9890793, 19767395 ,9987115, 11544224, 17981983, 14763982, 8016066 19361434 18689473 20070525 20923420 20852588 22517742 21622759 21368759 21821766 23595989 18786145,16525504,17380125, 16163393,12598648 24263382</pubmed> | + | <pubmed>9987115,8016067,9000055,12923101,10447896,9141693,2113920,16497325,19226326,8793870,10809708,14679237,17560370,11684022,8195092,11722737,11914365,12028382,18689476,19361434,9890793, 19767395 ,9987115, 11544224, 17981983, 14763982, 8016066 19361434 18689473 20070525 20923420 20852588 22517742 21622759 21368759 21821766 23595989 18786145,16525504,17380125, 16163393,12598648 24263382 25610436</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:05, 23 January 2015
- Description: ATPase subunit of the ATP-dependent ClpC-ClpP protease, involved in competence development, heat shock regulation, motility, sporulation, protein quality control, biofilm formation
| Gene name | clpC |
| Synonyms | mecB |
| Essential | no |
| Product | ATPase subunit of the ClpC-ClpP protease |
| Function | protein degradation positive regulator of autolysin (LytC and LytD) synthesis |
| Gene expression levels in SubtiExpress: clpC | |
| Interactions involving this protein in SubtInteract: ClpC | |
| Metabolic function and regulation of this protein in SubtiPathways: clpC | |
| MW, pI | 89 kDa, 5.746 |
| Gene length, protein length | 2430 bp, 810 aa |
| Immediate neighbours | mcsB, radA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 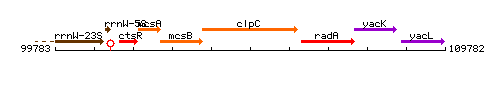
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
proteolysis, sporulation proteins, general stress proteins (controlled by SigB), heat shock proteins, phosphoproteins
This gene is a member of the following regulons
CtsR regulon, SigB regulon, SigF regulon
The gene
Basic information
- Locus tag: BSU00860
Phenotypes of a mutant
Database entries
- BsubCyc: BSU00860
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
- Protein family: mecA family (according to Swiss-Prot) clpA/clpB family. ClpC subfamily (according to Swiss-Prot), AAA+ -type ATPase (IPR013093) InterPro (PF07724) PFAM
Targets of ClpC-ClpP-dependent protein degradation
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-5 and Arg-254 PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU00860
- UniProt: P37571
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 1157 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 2624 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 711 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 495 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 617 PubMed
Biological materials
- Mutant:
- clpC::tet available from the Hamoen Lab
- BP98 (clpP::spc), available in Fabian Commichau's lab PubMed
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal GFP fusions (single copy, also as CFP and YFP variants) available from the Hamoen Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Leendert Hamoen, Newcastle University, UK homepage
- Kürsad Turgay, Freie Universität Berlin, Germany homepage
Your additional remarks
References
Reviews
Noël Molière, Kürşad Turgay
General and regulatory proteolysis in Bacillus subtilis.
Subcell Biochem: 2013, 66;73-103
[PubMed:23479438]
[WorldCat.org]
[DOI]
(P p)
Aurelia Battesti, Susan Gottesman
Roles of adaptor proteins in regulation of bacterial proteolysis.
Curr Opin Microbiol: 2013, 16(2);140-7
[PubMed:23375660]
[WorldCat.org]
[DOI]
(I p)
Noël Molière, Kürşad Turgay
Chaperone-protease systems in regulation and protein quality control in Bacillus subtilis.
Res Microbiol: 2009, 160(9);637-44
[PubMed:19781636]
[WorldCat.org]
[DOI]
(I p)
Janine Kirstein, Noël Molière, David A Dougan, Kürşad Turgay
Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases.
Nat Rev Microbiol: 2009, 7(8);589-99
[PubMed:19609260]
[WorldCat.org]
[DOI]
(I p)
Dorte Frees, Kirsi Savijoki, Pekka Varmanen, Hanne Ingmer
Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, Gram-positive bacteria.
Mol Microbiol: 2007, 63(5);1285-95
[PubMed:17302811]
[WorldCat.org]
[DOI]
(P p)
Original Publications