SubtiWiki v3 inherited the same user information as the previous versions. The same user account can be used to log in to any versions of SubtiWiki
Please contact web admin bzhu@gwdg.de to reset the password
Please contact admin jstuelke@gwdg.de to create an account
Please contact admin jstuelk@gwdg.de
SubtiWiki v3 has adapted a new markup system. This markup system not only indicate the style of the page, but also the structure the page. The text below is a part of source text of gene dnaA
* description: AAA ATPase, replication initiation protein
* locus: BSU00010
* pI: 6.03
* mw: 50.695
* proteinLength: 446
* Gene
** Coordinates: 410 → 1,750
** Phenotypes of a mutant
essential [Pubmed|12682299]
In this markup system, all section titles starts with a asterisk " * ", the primary section title with one " * " while the secondary section title with two " * ".
Content of the section can be placed either right behind the title, or under the title. A new line in the edit box corresponds to a bullet point when parsed.
A universal template is provided for gene's page in SubtiWiki. Information can be added to intended section.
Note:
A space is needed between " * " and the title, as well as between " : " and the content.
| Name | Syntax | Parsed | Hot key |
|---|---|---|---|
| gene | [[gene|dnaA]] [[gene|ptsH|Hpr]] |
[gene|6740108089F13116F200C15F35C2E7561E990FEB|dnaA] [gene|29B793660E4D30C0656248F3EF403FEF76FB9025|Hpr] |
ctrl + shift + g |
| protein | [[protein|dnaA]] [[protein|ptsH|Hpr]] |
[protein|6740108089F13116F200C15F35C2E7561E990FEB|DnaA] [protein|29B793660E4D30C0656248F3EF403FEF76FB9025|Hpr] |
ctrl + shift + p |
| SubtiWiki | [SW|Ribosome] | [SW|Ribosome] | ctrl + shift + a |
| PDB | [PDB|2Z4R] | [PDB|2Z4R] | None |
| PUBMED | [Pubmed|23909787,22286949] | [Pubmed|23909787,22286949] | ctrl + shift + c |
| REGULON | [[regulon|dnaA]] [[regulon|T-box]] |
[regulon|6740108089F13116F200C15F35C2E7561E990FEB|dnaA] [regulon|T-box|T-box] |
None |
| External url | [https://www.google.de/ Google] | [https://www.google.de/ Google] | None |
| I(i) | <i>italic text</i> | italic text | None |
| B(b) | <b>bold text</b> | bold text | None |
| X2(down) | X<sub>2</sub> | X2 | None |
| X2(up) | X<sup>2</sup> | X2 | None |
| Citation | <pubmed>23909787,22286949</pubmed> | Citation in green box | None |
In this version of SubtiWiki, the editing page of a gene provides portals of all editing interface concerning this gene.
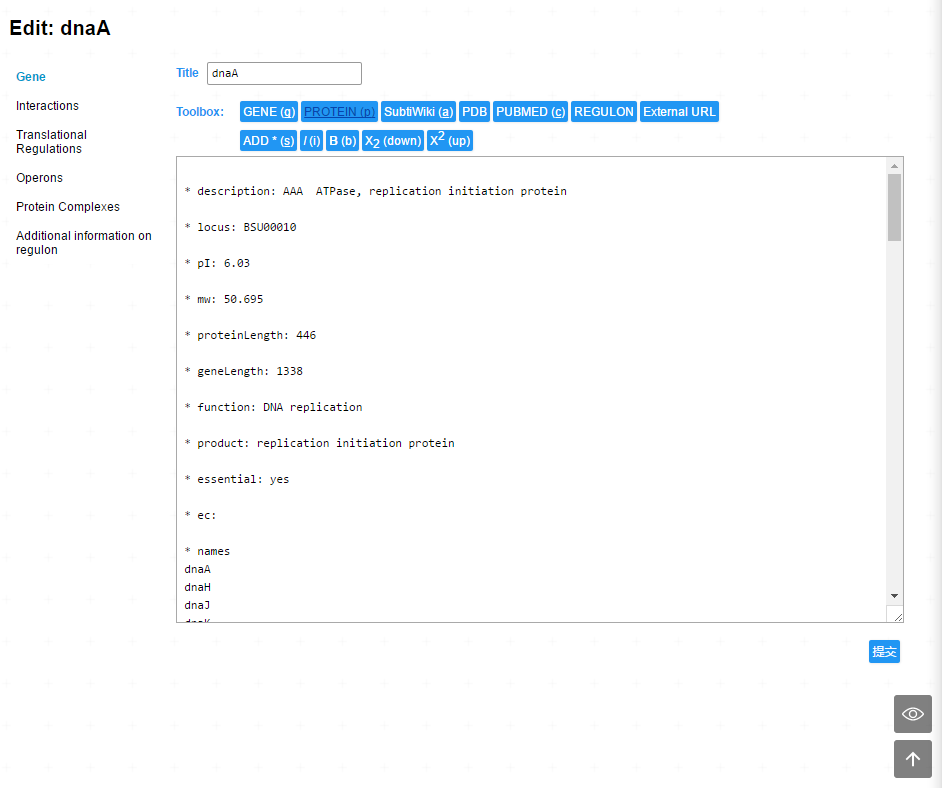
| Section name | Description |
|---|---|
| Gene | General information about this gene, transcribed RNA, and translated protein (if exists). |
| Interactions | Information of protein-protein interactions. |
| Translational regulations | regulation which happens at translational level, transcriptional regulation please go to "Operons" section |
| Operon | all operons this gene is in, transcriptional regulation data stored here. |
| Protein Complexes | functional protein complexes, still under construction, but you are welcome to add in data already. |
| Additional information on regulon | If this protein is a regulator, some information about this protein as regulation can be added here, such as the reference papers or etc. |
In the new regulation browser, a sub network centering the target gene is displayed. Radius is informally defined as 'how many steps it take from the target gene into the regulatory network'. Formally speaking, radius is the maximal distance between any gene in this sub network to target gene.
The zoom button in the settings panel controls the radius.
It is the ratio of genes in this sub network over all genes in B.subtilis subsp 168
Right click on the white area in the regulation browser. A pop up menu provides the link to export the data as csv, export the network as image, and export the network in NetVis supported format.
NetVis is a simple cross-platform network visualizer. It can be seen as the offline version of regulation browser. It uses the same visualization engine and from regulation browser network can be exported and viewed later locally.