SigF
- Description: RNA polymerase forespore-specific (early) sigma factor SigF
| Gene name | sigF |
| Synonyms | spoIIAC |
| Essential | no |
| Product | RNA polymerase forespore-specific (early) sigma factor SigF |
| Function | transcription of sporulation genes (late mother cell) |
| Interactions involving this protein in SubtInteract: SigF | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 29 kDa, 5.078 |
| Gene length, protein length | 765 bp, 255 aa |
| Immediate neighbours | spoVAA, spoIIAB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 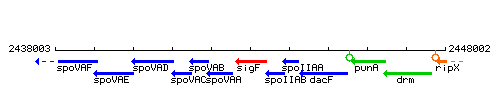
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, sporulation proteins, membrane proteins
This gene is a member of the following regulons
AbrB regulon, SigF regulon, SigG regulon, SigH regulon, SinR regulon, Spo0A regulon
The SigF regulon
The gene
Basic information
- Locus tag: BSU23450
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s): SigG
Genes controlled by SigF
dacF-spoIIAA-spoIIAB-sigF, sigG, pbpF, yphA-seaA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure: 1L0O (complex with SpoIIAB, Geobacillus stearothermophilus)
- UniProt: P07860
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Reviews
Michiel J L de Hoon, Patrick Eichenberger, Dennis Vitkup
Hierarchical evolution of the bacterial sporulation network.
Curr Biol: 2010, 20(17);R735-45
[PubMed:20833318]
[WorldCat.org]
[DOI]
(I p)
The SigF regulon
Original Publications
Additional publications: PubMed
Jin Hwan Do, Masao Nagasaki, Satoru Miyano
The systems approach to the prespore-specific activation of sigma factor SigF in Bacillus subtilis.
Biosystems: 2010, 100(3);178-84
[PubMed:20298743]
[WorldCat.org]
[DOI]
(I p)
Amy H Camp, Richard Losick
A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis.
Genes Dev: 2009, 23(8);1014-24
[PubMed:19390092]
[WorldCat.org]
[DOI]
(I p)
Céline Karmazyn-Campelli, Lamya Rhayat, Rut Carballido-López, Sandra Duperrier, Niels Frandsen, Patrick Stragier
How the early sporulation sigma factor sigmaF delays the switch to late development in Bacillus subtilis.
Mol Microbiol: 2008, 67(5);1169-80
[PubMed:18208527]
[WorldCat.org]
[DOI]
(I p)
Oleg A Igoshin, Margaret S Brody, Chester W Price, Michael A Savageau
Distinctive topologies of partner-switching signaling networks correlate with their physiological roles.
J Mol Biol: 2007, 369(5);1333-52
[PubMed:17498739]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, José Eduardo González-Pastor, Richard Losick
High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis.
J Bacteriol: 2005, 187(4);1357-68
[PubMed:15687200]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Tae-Jong Kim, Chester W Price, Richard Losick
Insulation of the sigmaF regulatory system in Bacillus subtilis.
J Bacteriol: 2004, 186(13);4390-4
[PubMed:15205443]
[WorldCat.org]
[DOI]
(P p)
Margaret S Ho, Karen Carniol, Richard Losick
Evidence in support of a docking model for the release of the transcription factor sigma F from the antisigma factor SpoIIAB in Bacillus subtilis.
J Biol Chem: 2003, 278(23);20898-905
[PubMed:12676949]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
Jennifer T Kemp, Adam Driks, Richard Losick
FtsA mutants of Bacillus subtilis impaired in sporulation.
J Bacteriol: 2002, 184(14);3856-63
[PubMed:12081956]
[WorldCat.org]
[DOI]
(P p)
Q Pan, D A Garsin, R Losick
Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis.
Mol Cell: 2001, 8(4);873-83
[PubMed:11684022]
[WorldCat.org]
[DOI]
(P p)
N Frandsen, I Barák, C Karmazyn-Campelli, P Stragier
Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis.
Genes Dev: 1999, 13(4);394-9
[PubMed:10049355]
[WorldCat.org]
[DOI]
(P p)
K Pogliano, A E Hofmeister, R Losick
Disappearance of the sigma E transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis.
J Bacteriol: 1997, 179(10);3331-41
[PubMed:9150232]
[WorldCat.org]
[DOI]
(P p)
M Gomez, S M Cutting
Expression of the Bacillus subtilis spoIVB gene is under dual sigma F/sigma G control.
Microbiology (Reading): 1996, 142 ( Pt 12);3453-7
[PubMed:9004507]
[WorldCat.org]
[DOI]
(P p)
A L Decatur, R Losick
Three sites of contact between the Bacillus subtilis transcription factor sigmaF and its antisigma factor SpoIIAB.
Genes Dev: 1996, 10(18);2348-58
[PubMed:8824593]
[WorldCat.org]
[DOI]
(P p)
A Feucht, T Magnin, M D Yudkin, J Errington
Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis.
Genes Dev: 1996, 10(7);794-803
[PubMed:8846916]
[WorldCat.org]
[DOI]
(P p)
C D Webb, A Decatur, A Teleman, R Losick
Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis.
J Bacteriol: 1995, 177(20);5906-11
[PubMed:7592342]
[WorldCat.org]
[DOI]
(P p)
R Schuch, P J Piggot
The dacF-spoIIA operon of Bacillus subtilis, encoding sigma F, is autoregulated.
J Bacteriol: 1994, 176(13);4104-10
[PubMed:8021191]
[WorldCat.org]
[DOI]
(P p)
S Alper, L Duncan, R Losick
An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis.
Cell: 1994, 77(2);195-205
[PubMed:8168129]
[WorldCat.org]
[DOI]
(P p)
D Foulger, J Errington
Effects of new mutations in the spoIIAB gene of Bacillus subtilis on the regulation of sigma F and sigma G activities.
J Gen Microbiol: 1993, 139(12);3197-203
[PubMed:8126438]
[WorldCat.org]
[DOI]
(P p)
J J Wu, R Schuch, P J Piggot
Characterization of a Bacillus subtilis sporulation operon that includes genes for an RNA polymerase sigma factor and for a putative DD-carboxypeptidase.
J Bacteriol: 1992, 174(15);4885-92
[PubMed:1629150]
[WorldCat.org]
[DOI]
(P p)
K York, T J Kenney, S Satola, C P Moran, H Poth, P Youngman
Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE.
J Bacteriol: 1992, 174(8);2648-58
[PubMed:1556084]
[WorldCat.org]
[DOI]
(P p)
P Margolis, A Driks, R Losick
Establishment of cell type by compartmentalized activation of a transcription factor.
Science: 1991, 254(5031);562-5
[PubMed:1948031]
[WorldCat.org]
[DOI]
(P p)
R Schmidt, P Margolis, L Duncan, R Coppolecchia, C P Moran, R Losick
Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1990, 87(23);9221-5
[PubMed:2123551]
[WorldCat.org]
[DOI]
(P p)
J Errington, J Mandelstam
Use of a lacZ gene fusion to determine the dependence pattern of sporulation operon spoIIA in spo mutants of Bacillus subtilis.
J Gen Microbiol: 1986, 132(11);2967-76
[PubMed:3114419]
[WorldCat.org]
[DOI]
(P p)