IolE
- Description: 2-keto-myo-inositol dehydratase, dehydration of 2-keto-myo-inositol (2nd reaction)
| Gene name | iolE |
| Synonyms | yxdE |
| Essential | no |
| Product | 2-keto-myo-inositol dehydratase, dehydration of 2-keto-myo-inositol (2nd reaction) |
| Function | myo-inositol catabolism |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 33 kDa, 5.209 |
| Gene length, protein length | 891 bp, 297 aa |
| Immediate neighbours | iolF, iolD |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 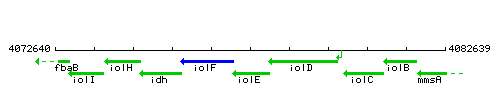
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU39720
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2,4,6/3,5-pentahydroxycyclohexanone = 3,5/4-trihydroxycyclohexa-1,2-dione + H2O (according to Swiss-Prot)
- Protein family: iolE/mocC family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P42416
- KEGG entry: [3]
- E.C. number: 4.2.1.44
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Yasutaro Fujita, University of Fukuyama, Japan
- Ken-ichi Yoshida, Kobe University, Japan
Your additional remarks
References