Sandbox
- Description: Carbon catabolite control protein A, involved in glucose regulation of many genes; represses catabolic genes and activates genes involved in excretion of excess carbon
| Gene name | ccpA |
| Synonyms | graR, alsA, amyR |
| Essential | no |
| Product | transcriptional regulator |
| Function | mediates carbon catabolite repression (CCR) |
| Metabolic function and regulation of this protein in SubtiPathways: Nucleoside catabolism, Nucleotides (regulation), Ile, Leu, Val, His, Coenzyme A, Central C-metabolism | |
| MW, pI | 36,8 kDa, 5.06 |
| Gene length, protein length | 1002 bp, 334 amino acids |
| Immediate neighbours | aroA, motP |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 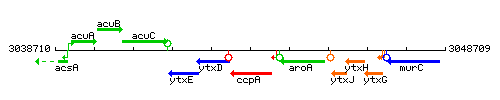
This image was kindly provided by SubtiList
| |
Contents
[hide]
The gene
Basic information
- Locus tag: BSU29740
Phenotypes of a mutant
Loss of carbon catabolite repression. Loss of PTS-dependent sugar transport due to excessive phosphorylation of HPr by HprK. The mutant is unable to grow on a minimal medium with glucose and ammonium as the only sources of carbon and nitrogen, respectively.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcriptional regulator of carbon catabolite repression (CCR)
- Protein family: LacI family
- Paralogous protein(s):
Genes controlled by CcpA
- Repression by CcpA: abbA, amyE, bglP-bglH, bglS, cccA, citZ-icd-mdh, levD-levE-levF-levG-sacC, licB-licC-licA-licH, phoP-phoR, xylA-xylB, xynP-xynB
Extended information on the protein
- Kinetic information:
- Domains:
- HTH lacI-type Domain (1 – 58)
- DNA binding Domain (6 – 25)
- Modification:
- Cofactor(s): HPr-Ser46-P, Crh-Ser-46-P
- Effectors of protein activity:glucose-6-phosphate, fructose-1,6-bisphosphate Pubmed
- Localization:
Database entries
- UniProt entry: P25144
- KEGG entry: [3]
Additional information
Expression and regulation
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Additional information: there are about 3.000 molecules of CcpA per cell PubMed
Biological materials
- Expression vector: pGP643 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
Labs working on this gene/protein
Wolfgang Hillen, Erlangen University, Germany Homepage
Richard Brennan, Houston, Texas, USA Homepage
Milton H. Saier, University of California at San Diego, USA Homepage
Yasutaro Fujita, University of Fukuyama, Japan
Jörg Stülke, University of Göttingen, Germany Homepage
Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
Reviews
General and physiological studies
Kalpana D Singh, Matthias H Schmalisch, Jörg Stülke, Boris Görke
Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources.
J Bacteriol: 2008, 190(21);7275-84
[PubMed:18757537]
[WorldCat.org]
[DOI]
(I p)
Naoya Terahara, Makoto Fujisawa, Benjamin Powers, Tina M Henkin, Terry A Krulwich, Masahiro Ito
An intergenic stem-loop mutation in the Bacillus subtilis ccpA-motPS operon increases motPS transcription and the MotPS contribution to motility.
J Bacteriol: 2006, 188(7);2701-5
[PubMed:16547058]
[WorldCat.org]
[DOI]
(P p)
Ingrid Wacker, Holger Ludwig, Irene Reif, Hans-Matti Blencke, Christian Detsch, Jörg Stülke
The regulatory link between carbon and nitrogen metabolism in Bacillus subtilis: regulation of the gltAB operon by the catabolite control protein CcpA.
Microbiology (Reading): 2003, 149(Pt 10);3001-3009
[PubMed:14523131]
[WorldCat.org]
[DOI]
(P p)
Holger Ludwig, Nicole Rebhan, Hans-Matti Blencke, Matthias Merzbacher, Jörg Stülke
Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation.
Mol Microbiol: 2002, 45(2);543-53
[PubMed:12123463]
[WorldCat.org]
[DOI]
(P p)
N Faires, S Tobisch, S Bachem, I Martin-Verstraete, M Hecker, J Stülke
The catabolite control protein CcpA controls ammonium assimilation in Bacillus subtilis.
J Mol Microbiol Biotechnol: 1999, 1(1);141-8
[PubMed:10941796]
[WorldCat.org]
(P p)
Y Miwa, M Saikawa, Y Fujita
Possible function and some properties of the CcpA protein of Bacillus subtilis.
Microbiology (Reading): 1994, 140 ( Pt 10);2567-75
[PubMed:8000527]
[WorldCat.org]
[DOI]
(P p)
T M Henkin, F J Grundy, W L Nicholson, G H Chambliss
Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors.
Mol Microbiol: 1991, 5(3);575-84
[PubMed:1904524]
[WorldCat.org]
[DOI]
(P p)
Global analyses (proteome, transcriptome)
Repression of target genes by CcpA
Positive regulation of gene expression by CcpA
Control of CcpA activity
CcpA-DNA interaction
Gerald Seidel, Marco Diel, Norbert Fuchsbauer, Wolfgang Hillen
Quantitative interdependence of coeffectors, CcpA and cre in carbon catabolite regulation of Bacillus subtilis.
FEBS J: 2005, 272(10);2566-77
[PubMed:15885105]
[WorldCat.org]
[DOI]
(P p)
Y Miwa, A Nakata, A Ogiwara, M Yamamoto, Y Fujita
Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis.
Nucleic Acids Res: 2000, 28(5);1206-10
[PubMed:10666464]
[WorldCat.org]
[DOI]
(I p)
J H Kim, G H Chambliss
Contacts between Bacillus subtilis catabolite regulatory protein CcpA and amyO target site.
Nucleic Acids Res: 1997, 25(17);3490-6
[PubMed:9254709]
[WorldCat.org]
[DOI]
(P p)
Y Fujita, Y Miwa, A Galinier, J Deutscher
Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr.
Mol Microbiol: 1995, 17(5);953-60
[PubMed:8596444]
[WorldCat.org]
[DOI]
(P p)
J H Kim, Z T Guvener, J Y Cho, K C Chung, G H Chambliss
Specificity of DNA binding activity of the Bacillus subtilis catabolite control protein CcpA.
J Bacteriol: 1995, 177(17);5129-34
[PubMed:7665492]
[WorldCat.org]
[DOI]
(P p)
Functional analysis of CcpA
H Ludwig, J Stülke
The Bacillus subtilis catabolite control protein CcpA exerts all its regulatory functions by DNA-binding.
FEMS Microbiol Lett: 2001, 203(1);125-9
[PubMed:11557150]
[WorldCat.org]
[DOI]
(P p)
E Küster-Schöck, A Wagner, U Völker, W Hillen
Mutations in catabolite control protein CcpA showing glucose-independent regulation in Bacillus megaterium.
J Bacteriol: 1999, 181(24);7634-8
[PubMed:10601226]
[WorldCat.org]
[DOI]
(P p)
E Küster, T Hilbich, M K Dahl, W Hillen
Mutations in catabolite control protein CcpA separating growth effects from catabolite repression.
J Bacteriol: 1999, 181(13);4125-8
[PubMed:10383986]
[WorldCat.org]
[DOI]
(P p)
A Kraus, E Küster, A Wagner, K Hoffmann, W Hillen
Identification of a co-repressor binding site in catabolite control protein CcpA.
Mol Microbiol: 1998, 30(5);955-63
[PubMed:9988473]
[WorldCat.org]
[DOI]
(P p)
A Kraus, W Hillen
Analysis of CcpA mutations defective in carbon catabolite repression in Bacillus megaterium.
FEMS Microbiol Lett: 1997, 153(1);221-6
[PubMed:9252590]
[WorldCat.org]
[DOI]
(P p)
Structural analyses
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose 6-phosphate and fructose 1,6-bisphosphate.
J Mol Biol: 2007, 368(4);1042-50
[PubMed:17376479]
[WorldCat.org]
[DOI]
(P p)
Maria A Schumacher, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.
J Biol Chem: 2006, 281(10);6793-800
[PubMed:16316990]
[WorldCat.org]
[DOI]
(P p)
Maria A Schumacher, Gregory S Allen, Marco Diel, Gerald Seidel, Wolfgang Hillen, Richard G Brennan
Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell: 2004, 118(6);731-41
[PubMed:15369672]
[WorldCat.org]
[DOI]
(P p)