Difference between revisions of "PnpA"
| Line 18: | Line 18: | ||
|style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2115 bp, 705 aa | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 2115 bp, 705 aa | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rpsO]]'', ''[[ylxY]]'' |
|- | |- | ||
|style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/pnpA_nucleotide.txt Gene sequence (+200bp) ]''' | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/pnpA_nucleotide.txt Gene sequence (+200bp) ]''' | ||
Revision as of 15:27, 20 February 2009
- Description: polynucleotide phosphorylase
| Gene name | pnpA |
| Synonyms | comR |
| Essential | no |
| Product | polynucleotide phosphorylase (PNPase) (EC 2.7.7.8) |
| Function | necessary for competence development |
| MW, pI | 77 kDa, 4.89 |
| Gene length, protein length | 2115 bp, 705 aa |
| Immediate neighbours | rpsO, ylxY |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 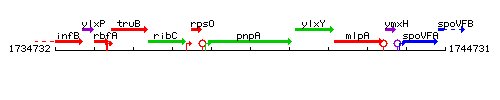
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 3'-5' exoribonuclease
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure: 3CDI (protein from E. coli)
- Swiss prot entry:
- KEGG entry:
- E.C. number:
Additional information
required for the expression of late competence genes comG and comK, requirement bypassed by a mecA disruption; may be necessary for modification of the srfA transcript (stabilization or translation activation)
Expression and regulation
- Operon:
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
David Bechhofer, Mount Sinai School, New York, USA Homepage
Your additional remarks
References
- Bechhofer, D. H. & Wang, W. (1998). Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J Bacteriol 180:5968-5977. PubMed
- Campos-Guillen, J., Bralley, P., Jones, G. H., Bechhofer, D. H. & Olmedo-Alvarez, G. (2005). Addition of poly(A) and heteropolymeric 3´ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J Bacteriol 187:4698-4706. PubMed
- Deutscher, M. P. & Reuven, N. B. (1991). Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci U S A 88:3277-3280. PubMed
- Mitra, S., Hue, K. & Bechhofer, D. H. (1996). In vitro processing activity of Bacillus subtilis polynucleotide phosphorylase. Mol Microbiol 19:329-342. PubMed
- Wang, W. & Bechhofer, D. H. (1996). Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol 178:2375-2382. PubMed
- Oussenko, I. A., Abe, T., Ujiie, H., Muto, A. & Bechhofer, D. H. (2005). Participation of 3'-to-5' exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol 187:2758-2767. PubMed
- Commichau, F. M., Rothe, F. M., Herzberg, C., Wagner, E., Hellwig, D., Lehnik-Habrink, M., Hammer, E., Völker, U. & Stülke, J. (2009) Novel activities of glycolytic enzymes in Bacillus subtilis: Interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics in press PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed