Difference between revisions of "SigM"
(→References) |
(→Phenotypes of a mutant) |
||
| Line 57: | Line 57: | ||
* ''sigM'' mutants form aberrantly shaped cells, which swell and lyse spontaneously during growth in NB medium containing increased levels (0.35-0.7 M) of a wide range of different salts {{PubMed|10216858}} | * ''sigM'' mutants form aberrantly shaped cells, which swell and lyse spontaneously during growth in NB medium containing increased levels (0.35-0.7 M) of a wide range of different salts {{PubMed|10216858}} | ||
* increased sensitivity towards beta-lactam antibiotics {{PubMed|22211522}} | * increased sensitivity towards beta-lactam antibiotics {{PubMed|22211522}} | ||
| + | * a ''[[sigM]] [[murJ]]'' double mutant is not viable (due to the lack of both lipid II flippases) {{PubMed|25918422}} | ||
=== Database entries === | === Database entries === | ||
Revision as of 13:00, 8 May 2015
- Description: RNA polymerase ECF-type sigma factor SigM
| Gene name | sigM |
| Synonyms | yhdM |
| Essential | no |
| Product | RNA polymerase ECF-type sigma factor SigM |
| Function | responsible for intrinsic resistance against beta-lactam antibiotics |
| Gene expression levels in SubtiExpress: sigM | |
| Interactions involving this protein in SubtInteract: SigM | |
| MW, pI | 19 kDa, 6.77 |
| Gene length, protein length | 489 bp, 163 aa |
| Immediate neighbours | yhdL, yhdN |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 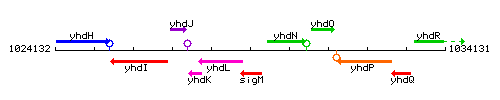
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against toxins/ antibiotics
This gene is a member of the following regulons
The SigM regulon
The gene
Basic information
- Locus tag: BSU09520
Phenotypes of a mutant
- SigM is essential for growth and survival in nutrient broth (NB) containing 1.4 M NaCl PubMed
- sigM mutants form aberrantly shaped cells, which swell and lyse spontaneously during growth in NB medium containing increased levels (0.35-0.7 M) of a wide range of different salts PubMed
- increased sensitivity towards beta-lactam antibiotics PubMed
- a sigM murJ double mutant is not viable (due to the lack of both lipid II flippases) PubMed
Database entries
- BsubCyc: BSU09520
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family, ECF subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU09520
- Structure:
- UniProt: O07582
- KEGG entry: [3]
- E.C. number:
Additional information
- Expression of the SigM regulon in increased in ugtP mutants PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- 1A906 (sigM::kan), PubMed, available at BGSC
- BP129 (sigM::aphA3), available in Fabian Commichau's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
Reviews
The SigM regulon
Other original publications