Difference between revisions of "RnjA"
(→Expression and regulation) |
(→Reviews) |
||
| Line 175: | Line 175: | ||
=References= | =References= | ||
==Reviews== | ==Reviews== | ||
| − | <pubmed>20458164 21334965 21893280 23403287,21957024 22568516 24064983 </pubmed> | + | <pubmed>20458164 21334965 21893280 23403287,21957024 22568516 24064983 25292357</pubmed> |
==Original publications== | ==Original publications== | ||
<pubmed>18079181, 19553197, 17981983, 19458035, 15831787, 18204464, 18713320, 19193632, 17005971, 18445592, 17512403, 17229210, 17576666, 19210617 19633085 19638340 19850915 19880604 20025672 20418391 20572937 ,21803996 21862575 21925382 22198292 22412379 23504012 21893285,21893286,21908660 24187087</pubmed> | <pubmed>18079181, 19553197, 17981983, 19458035, 15831787, 18204464, 18713320, 19193632, 17005971, 18445592, 17512403, 17229210, 17576666, 19210617 19633085 19638340 19850915 19880604 20025672 20418391 20572937 ,21803996 21862575 21925382 22198292 22412379 23504012 21893285,21893286,21908660 24187087</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:28, 8 March 2015
- Description: RNase J1
| Gene name | rnjA |
| Synonyms | ykqC |
| Essential | yes PubMed |
| Product | RNase J1 |
| Function | RNA processing |
| Gene expression levels in SubtiExpress: rnjA | |
| Interactions involving this protein in SubtInteract: RNase J1 | |
| Metabolic function and regulation of this protein in SubtiPathways: rnjA | |
| MW, pI | 61 kDa, 5.902 |
| Gene length, protein length | 1665 bp, 555 aa |
| Immediate neighbours | adeC, rpoY |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 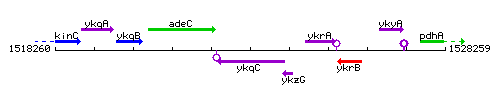
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14530
Phenotypes of a mutant
- essential PubMed
- a study from the lab of Ciaran Condon reports that rnjA is non-essential and that the mutant is strongly impaired in sporulation, genetic competence and many other phenotypes PubMed
Database entries
- BsubCyc: BSU14530
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: endonuclease and 5'-3' exonuclease
- Protein family: RNase J subfamily (according to Swiss-Prot)
- Paralogous protein(s): RnjB
RNAs affected by rnjA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU14530
- UniProt: Q45493
- KEGG entry: [2]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- required for thrS RNA processing, involved in maturation of the 5’-end of the16S rRNA
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
- translation of YkzG and RnjA is coupled, and this coupling is required for efficient expression of RNase J1 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 2868 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 4928 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 2768 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 4125 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 5056 PubMed
Biological materials
- Mutant:
- GP41 (rnjA under control of p(xyl)), available in Jörg Stülke's lab
- SSB342 (rnjA under pspac), cat, available in Harald Putzer lab
- Expression vector:
- for chromosomal expression of RNase J1-Strep (spc): GP1034, available in Jörg Stülke's lab
- for chromosomal expression of RNase J1-Strep (cat): GP1042, available in Jörg Stülke's lab
- lacZ fusion: pGP418 (in pAC7), available in Jörg Stülke's lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1020 (spc, based on pGP1331), available in Jörg Stülke's lab
- GP1075 (aphA3), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Harald Putzer, IBPC Paris, France Homepage
David Bechhofer, Mount Sinai School, New York, USA Homepage
Ciaran Condon, IBPC, Paris, France Homepage
Your additional remarks
References
Reviews
Monica P Hui, Patricia L Foley, Joel G Belasco
Messenger RNA degradation in bacterial cells.
Annu Rev Genet: 2014, 48;537-59
[PubMed:25292357]
[WorldCat.org]
[DOI]
(I p)
Soumaya Laalami, Léna Zig, Harald Putzer
Initiation of mRNA decay in bacteria.
Cell Mol Life Sci: 2014, 71(10);1799-828
[PubMed:24064983]
[WorldCat.org]
[DOI]
(I p)
Zbigniew Dominski, Agamemnon J Carpousis, Béatrice Clouet-d'Orval
Emergence of the β-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation.
Biochim Biophys Acta: 2013, 1829(6-7);532-51
[PubMed:23403287]
[WorldCat.org]
[DOI]
(P p)
Martin Lehnik-Habrink, Richard J Lewis, Ulrike Mäder, Jörg Stülke
RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases.
Mol Microbiol: 2012, 84(6);1005-17
[PubMed:22568516]
[WorldCat.org]
[DOI]
(I p)
David H Bechhofer
Bacillus subtilis mRNA decay: new parts in the toolkit.
Wiley Interdiscip Rev RNA: 2011, 2(3);387-94
[PubMed:21957024]
[WorldCat.org]
[DOI]
(I p)
Jamie Richards, Joel G Belasco
Ribonuclease J: how to lead a double life.
Structure: 2011, 19(9);1201-3
[PubMed:21893280]
[WorldCat.org]
[DOI]
(I p)
Ciarán Condon, David H Bechhofer
Regulated RNA stability in the Gram positives.
Curr Opin Microbiol: 2011, 14(2);148-54
[PubMed:21334965]
[WorldCat.org]
[DOI]
(I p)
Ciarán Condon
What is the role of RNase J in mRNA turnover?
RNA Biol: 2010, 7(3);316-21
[PubMed:20458164]
[WorldCat.org]
[DOI]
(I p)
Original publications