Difference between revisions of "ClpP"
(→Original Publications) |
|||
| Line 172: | Line 172: | ||
<pubmed> 16211032 17302811 23375660 23479438,19609260,19781636</pubmed> | <pubmed> 16211032 17302811 23375660 23479438,19609260,19781636</pubmed> | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>9643546,10809708,11807061,9987115,14679237,18689476,15317791,17586624,11684022,12923101,17560370, 16899079,19226326 , 20070525,9987115,11544224 14763982 9643546, 19767395 11112444 9535081 18689473 20305655 20852588 16200071 21969594 22080375 22517742 17380125,12598648,9890793,20049702,20049702 23927726 24263382 24226776 24417481 15378759 23361916 24942655 </pubmed> | + | <pubmed>9643546,10809708,11807061,9987115,14679237,18689476,15317791,17586624,11684022,12923101,17560370, 16899079,19226326 , 20070525,9987115,11544224 14763982 9643546, 19767395 11112444 9535081 18689473 20305655 20852588 16200071 21969594 22080375 22517742 17380125,12598648,9890793,20049702,20049702 23927726 24263382 24226776 24417481 15378759 23361916 24942655 25212124 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:44, 19 September 2014
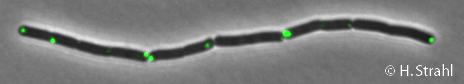
- Description: ATP-dependent Clp protease proteolytic subunit (class III heat-shock protein)
| Gene name | clpP |
| Synonyms | yvdN |
| Essential | no |
| Product | ATP-dependent Clp protease proteolytic subunit |
| Function | protein degradation |
| Gene expression levels in SubtiExpress: clpP | |
| Interactions involving this protein in SubtInteract: ClpP | |
| Metabolic function and regulation of this protein in SubtiPathways: clpP | |
| MW, pI | 21 kDa, 5.008 |
| Gene length, protein length | 591 bp, 197 aa |
| Immediate neighbours | trnQ-Arg, pgcM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 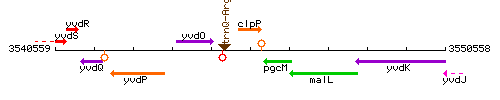
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed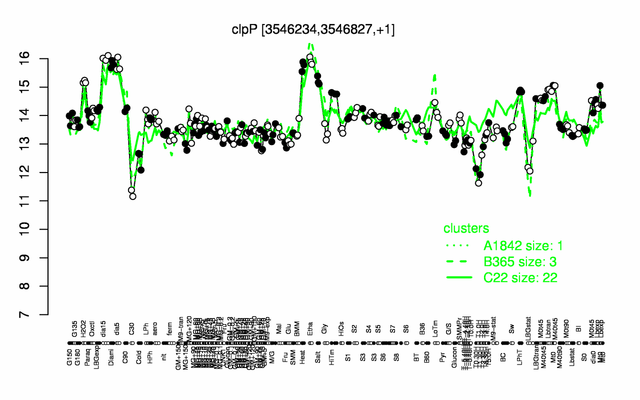
| |
Contents
Categories containing this gene/protein
proteolysis, general stress proteins (controlled by SigB), heat shock proteins, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU34540
Phenotypes of a mutant
- increased thermotolerance due to increased stabiliy of Spx and thus increased expression of trxA PubMed
Database entries
- BsubCyc: BSU34540
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of proteins to small peptides in the presence of ATP and magnesium (according to Swiss-Prot) endopeptidase/proteolysis
- Protein family: peptidase S14 family (according to Swiss-Prot) ClpP (IPR001907) InterPro, (PF00574) PFAM
- Paralogous protein(s):
Targets of ClpC-ClpP-dependent protein degradation
Targets of ClpX-ClpP-dependent protein degradation
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-13 PubMed
- Effectors of protein activity:
Database entries
- BsubCyc: BSU34540
- UniProt: P80244
- KEGG entry: [3]
- E.C. number: 3.4.21.92
Additional information
Expression and regulation
- Operon: clpP PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 4011 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 11118 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 1536 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 731 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1056 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal GFP fusions (both single copy and as 2th copy in amyE locus, also as CFP and YFP variants) available in the Leendert Hamoen lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Your additional remarks
References
Reviews
Original Publications
Daniel W Carney, Corey L Compton, Karl R Schmitz, Julia P Stevens, Robert T Sauer, Jason K Sello
A simple fragment of cyclic acyldepsipeptides is necessary and sufficient for ClpP activation and antibacterial activity.
Chembiochem: 2014, 15(15);2216-20
[PubMed:25212124]
[WorldCat.org]
[DOI]
(I p)
Chio Mui Chan, Erik Hahn, Peter Zuber
Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP.
Mol Microbiol: 2014, 93(3);426-38
[PubMed:24942655]
[WorldCat.org]
[DOI]
(I p)
Stephanie Runde, Noël Molière, Anja Heinz, Etienne Maisonneuve, Armgard Janczikowski, Alexander K W Elsholz, Ulf Gerth, Michael Hecker, Kürşad Turgay
The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis.
Mol Microbiol: 2014, 91(5);1036-52
[PubMed:24417481]
[WorldCat.org]
[DOI]
(I p)
Andreas Schmidt, Débora Broch Trentini, Silvia Spiess, Jakob Fuhrmann, Gustav Ammerer, Karl Mechtler, Tim Clausen
Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response.
Mol Cell Proteomics: 2014, 13(2);537-50
[PubMed:24263382]
[WorldCat.org]
[DOI]
(I p)
B P Conlon, E S Nakayasu, L E Fleck, M D LaFleur, V M Isabella, K Coleman, S N Leonard, R D Smith, J N Adkins, K Lewis
Activated ClpP kills persisters and eradicates a chronic biofilm infection.
Nature: 2013, 503(7476);365-70
[PubMed:24226776]
[WorldCat.org]
[DOI]
(I p)
John Alexopoulos, Bilal Ahsan, Lopamudra Homchaudhuri, Nabiha Husain, Yi-Qiang Cheng, Joaquin Ortega
Structural determinants stabilizing the axial channel of ClpP for substrate translocation.
Mol Microbiol: 2013, 90(1);167-80
[PubMed:23927726]
[WorldCat.org]
[DOI]
(I p)
Malte Gersch, Felix Gut, Vadim S Korotkov, Johannes Lehmann, Thomas Böttcher, Marion Rusch, Christian Hedberg, Herbert Waldmann, Gerhard Klebe, Stephan A Sieber
The mechanism of caseinolytic protease (ClpP) inhibition.
Angew Chem Int Ed Engl: 2013, 52(10);3009-14
[PubMed:23361916]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Byung-Gil Lee, Min Kyung Kim, Hyun Kyu Song
Structural insights into the conformational diversity of ClpP from Bacillus subtilis.
Mol Cells: 2011, 32(6);589-95
[PubMed:22080375]
[WorldCat.org]
[DOI]
(I p)
Peter Sass, Michaele Josten, Kirsten Famulla, Guido Schiffer, Hans-Georg Sahl, Leendert Hamoen, Heike Brötz-Oesterhelt
Antibiotic acyldepsipeptides activate ClpP peptidase to degrade the cell division protein FtsZ.
Proc Natl Acad Sci U S A: 2011, 108(42);17474-9
[PubMed:21969594]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Stephan Michalik, Daniela Zühlke, Michael Hecker, Ulf Gerth
CtsR, the Gram-positive master regulator of protein quality control, feels the heat.
EMBO J: 2010, 29(21);3621-9
[PubMed:20852588]
[WorldCat.org]
[DOI]
(I p)
Byung-Gil Lee, Eun Young Park, Kyung-Eun Lee, Hyesung Jeon, Kwang Hoon Sung, Holger Paulsen, Helga Rübsamen-Schaeff, Heike Brötz-Oesterhelt, Hyun Kyu Song
Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism.
Nat Struct Mol Biol: 2010, 17(4);471-8
[PubMed:20305655]
[WorldCat.org]
[DOI]
(I p)
Mitsuo Ogura, Kensuke Tsukahara
Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP.
Mol Microbiol: 2010, 75(5);1244-59
[PubMed:20070525]
[WorldCat.org]
[DOI]
(I p)
Janine Kirstein, Anja Hoffmann, Hauke Lilie, Ronny Schmidt, Helga Rübsamen-Waigmann, Heike Brötz-Oesterhelt, Axel Mogk, Kürşad Turgay
The antibiotic ADEP reprogrammes ClpP, switching it from a regulated to an uncontrolled protease.
EMBO Mol Med: 2009, 1(1);37-49
[PubMed:20049702]
[WorldCat.org]
[DOI]
(I p)
Ziqing Mei, Feng Wang, Yutao Qi, Zhiyuan Zhou, Qi Hu, Han Li, Jiawei Wu, Yigong Shi
Molecular determinants of MecA as a degradation tag for the ClpCP protease.
J Biol Chem: 2009, 284(49);34366-75
[PubMed:19767395]
[WorldCat.org]
[DOI]
(I p)
Jeanette Hahn, Naomi Kramer, Kenneth Briley, David Dubnau
McsA and B mediate the delocalization of competence proteins from the cell poles of Bacillus subtilis.
Mol Microbiol: 2009, 72(1);202-15
[PubMed:19226326]
[WorldCat.org]
[DOI]
(I p)
James Kain, Gina G He, Richard Losick
Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis.
J Bacteriol: 2008, 190(20);6749-57
[PubMed:18689476]
[WorldCat.org]
[DOI]
(I p)
Lyle A Simmons, Alan D Grossman, Graham C Walker
Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis.
J Bacteriol: 2008, 190(20);6758-68
[PubMed:18689473]
[WorldCat.org]
[DOI]
(I p)
Adam Reeves, Ulf Gerth, Uwe Völker, W G Haldenwang
ClpP modulates the activity of the Bacillus subtilis stress response transcription factor, sigmaB.
J Bacteriol: 2007, 189(17);6168-75
[PubMed:17586624]
[WorldCat.org]
[DOI]
(P p)
Peter Prepiak, David Dubnau
A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP.
Mol Cell: 2007, 26(5);639-47
[PubMed:17560370]
[WorldCat.org]
[DOI]
(P p)
Janine Kirstein, David A Dougan, Ulf Gerth, Michael Hecker, Kürşad Turgay
The tyrosine kinase McsB is a regulated adaptor protein for ClpCP.
EMBO J: 2007, 26(8);2061-70
[PubMed:17380125]
[WorldCat.org]
[DOI]
(P p)
Stephan Zellmeier, Wolfgang Schumann, Thomas Wiegert
Involvement of Clp protease activity in modulating the Bacillus subtilissigmaw stress response.
Mol Microbiol: 2006, 61(6);1569-82
[PubMed:16899079]
[WorldCat.org]
[DOI]
(P p)
Heike Brötz-Oesterhelt, Dieter Beyer, Hein-Peter Kroll, Rainer Endermann, Christoph Ladel, Werner Schroeder, Berthold Hinzen, Siegfried Raddatz, Holger Paulsen, Kerstin Henninger, Julia E Bandow, Hans-Georg Sahl, Harald Labischinski
Dysregulation of bacterial proteolytic machinery by a new class of antibiotics.
Nat Med: 2005, 11(10);1082-7
[PubMed:16200071]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Holger Kock, Ulf Gerth, Michael Hecker
The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis.
J Bacteriol: 2004, 186(17);5856-64
[PubMed:15317791]
[WorldCat.org]
[DOI]
(P p)
Holger Kock, Ulf Gerth, Michael Hecker
MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis.
Mol Microbiol: 2004, 51(4);1087-102
[PubMed:14763982]
[WorldCat.org]
[DOI]
(P p)
Ulf Gerth, Janine Kirstein, Jörg Mostertz, Torsten Waldminghaus, Marcus Miethke, Holger Kock, Michael Hecker
Fine-tuning in regulation of Clp protein content in Bacillus subtilis.
J Bacteriol: 2004, 186(1);179-91
[PubMed:14679237]
[WorldCat.org]
[DOI]
(P p)
Qi Pan, Richard Losick
Unique degradation signal for ClpCP in Bacillus subtilis.
J Bacteriol: 2003, 185(17);5275-8
[PubMed:12923101]
[WorldCat.org]
[DOI]
(P p)
Tilman Schlothauer, Axel Mogk, David A Dougan, Bernd Bukau, Kürşad Turgay
MecA, an adaptor protein necessary for ClpC chaperone activity.
Proc Natl Acad Sci U S A: 2003, 100(5);2306-11
[PubMed:12598648]
[WorldCat.org]
[DOI]
(P p)
Tiina Pummi, Soile Leskelä, Eva Wahlström, Ulf Gerth, Harold Tjalsma, Michael Hecker, Matti Sarvas, Vesa P Kontinen
ClpXP protease regulates the signal peptide cleavage of secretory preproteins in Bacillus subtilis with a mechanism distinct from that of the Ecs ABC transporter.
J Bacteriol: 2002, 184(4);1010-8
[PubMed:11807061]
[WorldCat.org]
[DOI]
(P p)
Q Pan, D A Garsin, R Losick
Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-sigma factor in B. subtilis.
Mol Cell: 2001, 8(4);873-83
[PubMed:11684022]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
H Nanamiya, K Takahashi, M Fujita, F Kawamura
Deficiency of the initiation events of sporulation in Bacillus subtilis clpP mutant can be suppressed by a lack of the Spo0E protein phosphatase.
Biochem Biophys Res Commun: 2000, 279(1);229-33
[PubMed:11112444]
[WorldCat.org]
[DOI]
(P p)
E Krüger, E Witt, S Ohlmeier, R Hanschke, M Hecker
The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins.
J Bacteriol: 2000, 182(11);3259-65
[PubMed:10809708]
[WorldCat.org]
[DOI]
(P p)
I Derré, G Rapoport, T Msadek
CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria.
Mol Microbiol: 1999, 31(1);117-31
[PubMed:9987115]
[WorldCat.org]
[DOI]
(P p)
K Turgay, J Hahn, J Burghoorn, D Dubnau
Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor.
EMBO J: 1998, 17(22);6730-8
[PubMed:9890793]
[WorldCat.org]
[DOI]
(P p)
U Gerth, E Krüger, I Derré, T Msadek, M Hecker
Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance.
Mol Microbiol: 1998, 28(4);787-802
[PubMed:9643546]
[WorldCat.org]
[DOI]
(P p)
T Msadek, V Dartois, F Kunst, M L Herbaud, F Denizot, G Rapoport
ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation.
Mol Microbiol: 1998, 27(5);899-914
[PubMed:9535081]
[WorldCat.org]
[DOI]
(P p)