Difference between revisions of "FlgN"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' flagellar filament assembly protein <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || flagellar filament assembly protein |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || flagellar filament assembly |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU35420 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU35420 flgN] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein= | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=FlgN FlgN] |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 18 kDa, 8.81 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 18 kDa, 8.81 | ||
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 59: | Line 55: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * non-motile cells {{PubMed|24706744}} | ||
=== Database entries === | === Database entries === | ||
| Line 68: | Line 65: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 86: | Line 80: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| Line 92: | Line 86: | ||
** phosphorylation on Tyr-49 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | ** phosphorylation on Tyr-49 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 115: | Line 109: | ||
=Expression and regulation= | =Expression and regulation= | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | ** ''[[comFA]]-[[comFB]]-[[comFC]]-[[yvyF]]-[[flgM]]-[[ | + | ** ''[[comFA]]-[[comFB]]-[[comFC]]-[[yvyF]]-[[flgM]]-[[flgN]]-[[flgK]]-[[flgL]]'' {{PubMed|8412657}} |
| − | ** ''[[yvyF]]-[[flgM]]-[[ | + | ** ''[[yvyF]]-[[flgM]]-[[flgN]]-[[flgK]]-[[flgL]]'' {{PubMed|8045879}} |
| − | ** ''[[flgM]]-[[ | + | ** ''[[flgM]]-[[flgN]]-[[flgK]]-[[flgL]]'' {{PubMed|21736639}} |
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yvyG_3639787_3640269_-1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=yvyG_3639787_3640269_-1 flgN] {{PubMed|22383849}} |
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
** ''[[comFA]]'': [[SigA]] {{PubMed|8412657}} | ** ''[[comFA]]'': [[SigA]] {{PubMed|8412657}} | ||
** ''[[yvyF]]'': [[SigD]] {{PubMed|8045879}} | ** ''[[yvyF]]'': [[SigD]] {{PubMed|8045879}} | ||
| Line 153: | Line 147: | ||
=References= | =References= | ||
| − | + | <pubmed>8412657 8045879 17218307, 15817382 22517742 24706744 21736639</pubmed> | |
| − | <pubmed>8412657 8045879 17218307, 15817382 22517742</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:33, 8 April 2014
- Description: flagellar filament assembly protein
| Gene name | yvyG |
| Synonyms | yviC |
| Essential | no |
| Product | flagellar filament assembly protein |
| Function | flagellar filament assembly |
| Gene expression levels in SubtiExpress: flgN | |
| Interactions involving this protein in SubtInteract: FlgN | |
| MW, pI | 18 kDa, 8.81 |
| Gene length, protein length | 480 bp, 160 aa |
| Immediate neighbours | flgK, flgM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 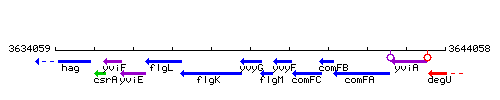
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, phosphoproteins
This gene is a member of the following regulons
ComK regulon, DegU regulon, SigD regulon
The gene
Basic information
- Locus tag: BSU35420
Phenotypes of a mutant
- non-motile cells PubMed
Database entries
- BsubCyc: BSU35420
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU35420
- Structure:
- UniProt: P39808
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Lynne S Cairns, Victoria L Marlow, Taryn B Kiley, Christopher Birchall, Adam Ostrowski, Phillip D Aldridge, Nicola R Stanley-Wall
FlgN is required for flagellum-based motility by Bacillus subtilis.
J Bacteriol: 2014, 196(12);2216-26
[PubMed:24706744]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Yi-Huang Hsueh, Loralyn M Cozy, Lok-To Sham, Rebecca A Calvo, Alina D Gutu, Malcolm E Winkler, Daniel B Kearns
DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis.
Mol Microbiol: 2011, 81(4);1092-108
[PubMed:21736639]
[WorldCat.org]
[DOI]
(I p)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Mark J Pallen, Charles W Penn, Roy R Chaudhuri
Bacterial flagellar diversity in the post-genomic era.
Trends Microbiol: 2005, 13(4);143-9
[PubMed:15817382]
[WorldCat.org]
[DOI]
(P p)
D B Mirel, P Lauer, M J Chamberlin
Identification of flagellar synthesis regulatory and structural genes in a sigma D-dependent operon of Bacillus subtilis.
J Bacteriol: 1994, 176(15);4492-500
[PubMed:8045879]
[WorldCat.org]
[DOI]
(P p)
J A Londoño-Vallejo, D Dubnau
comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases.
Mol Microbiol: 1993, 9(1);119-31
[PubMed:8412657]
[WorldCat.org]
[DOI]
(P p)