Difference between revisions of "PupG"
| Line 55: | Line 55: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23490&redirect=T BSU23490] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/drm-punA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/drm-punA.html] | ||
| Line 90: | Line 91: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU23490&redirect=T BSU23490] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 14:07, 2 April 2014
- Description: purine nucleoside phosphorylase
| Gene name | pupG |
| Synonyms | punA, pnp, yqkO |
| Essential | no |
| Product | purine nucleoside phosphorylase |
| Function | purine salvage and interconversion |
| Gene expression levels in SubtiExpress: pupG | |
| Metabolic function and regulation of this protein in SubtiPathways: pupG | |
| MW, pI | 28 kDa, 4.86 |
| Gene length, protein length | 813 bp, 271 aa |
| Immediate neighbours | dacF, drm |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 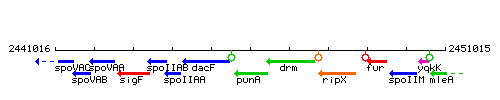
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed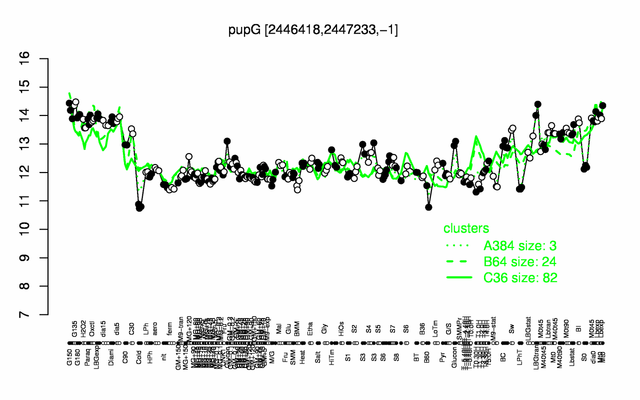
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of nucleotides, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU23490
Phenotypes of a mutant
Database entries
- BsubCyc: BSU23490
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Purine nucleoside + phosphate = purine + alpha-D-ribose 1-phosphate (according to Swiss-Prot) RNA(n+1) + phosphate = RNA(n) + a nucleoside diphosphate (according to Swiss-Prot)
- Protein family: PNP/MTAP phosphorylase family (according to Swiss-Prot) polyribonucleotide nucleotidyltransferase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-28 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Localization: secreted (according to Swiss-Prot)
Database entries
- BsubCyc: BSU23490
- Structure:
- UniProt: P46354
- KEGG entry: [3]
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
R Schuch, A Garibian, H H Saxild, P J Piggot, P Nygaard
Nucleosides as a carbon source in Bacillus subtilis: characterization of the drm-pupG operon.
Microbiology (Reading): 1999, 145 ( Pt 10);2957-66
[PubMed:10537218]
[WorldCat.org]
[DOI]
(P p)