Difference between revisions of "ComFA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' membrane-associated ATPase, may provide energy for DNA transport<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 39: | Line 39: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 84: | Line 80: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
** phosphorylated on Arg-20, Arg-186, and Arg-446 {{PubMed|22517742}} | ** phosphorylated on Arg-20, Arg-186, and Arg-446 {{PubMed|22517742}} | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 123: | Line 119: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=comFA_3642167_3643558_-1 comFA] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=comFA_3642167_3643558_-1 comFA] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' ''[[comFA]]'': [[SigA]] {{PubMed|8412657}} | + | * '''[[Sigma factor]]:''' ''[[comFA]]'': [[SigA]] {{PubMed|8412657}} |
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 11:17, 16 March 2014
- Description: membrane-associated ATPase, may provide energy for DNA transport
| Gene name | comFA |
| Synonyms | |
| Essential | no |
| Product | ATP-binding protein |
| Function | genetic competence |
| Gene expression levels in SubtiExpress: comFA | |
| Interactions involving this protein in SubtInteract: ComFA | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 52 kDa, 9.824 |
| Gene length, protein length | 1389 bp, 463 aa |
| Immediate neighbours | comFB, yviA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 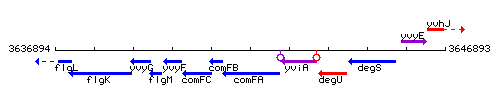
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
genetic competence, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35470
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: helicase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- phosphorylated on Arg-20, Arg-186, and Arg-446 PubMed
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39145
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Inês Chen, David Dubnau
DNA uptake during bacterial transformation.
Nat Rev Microbiol: 2004, 2(3);241-9
[PubMed:15083159]
[WorldCat.org]
[DOI]
(P p)
Original publications
Additional publications: PubMed