Difference between revisions of "YaaH"
(→Extended information on the protein) |
|||
| Line 77: | Line 77: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
| + | ** contains a N-acetylglucosamine-polymer-binding [[LysM domain]] {{PubMed|18430080}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 140: | Line 141: | ||
=References= | =References= | ||
| − | + | == Reviews== | |
| + | <pubmed>18430080</pubmed> | ||
| + | == Original publications == | ||
<pubmed>19087206,12884008,10419957,15805528 11964120, 19933362 22171814 15383836 22343356 18835992 </pubmed> | <pubmed>19087206,12884008,10419957,15805528 11964120, 19933362 22171814 15383836 22343356 18835992 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:04, 27 December 2013
- Description: general stress protein, survival of ethanol stress, SafA-dependent protein of the inner spore coat, spore cortex lytic protein
| Gene name | yaaH |
| Synonyms | sleL |
| Essential | no |
| Product | N-acetylglucosaminidase |
| Function | survival of ethanol stress, protection of the spore |
| Gene expression levels in SubtiExpress: yaaH | |
| MW, pI | 48 kDa, 5.618 |
| Gene length, protein length | 1281 bp, 427 aa |
| Immediate neighbours | dgk, yaaI |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 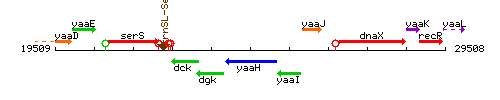
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed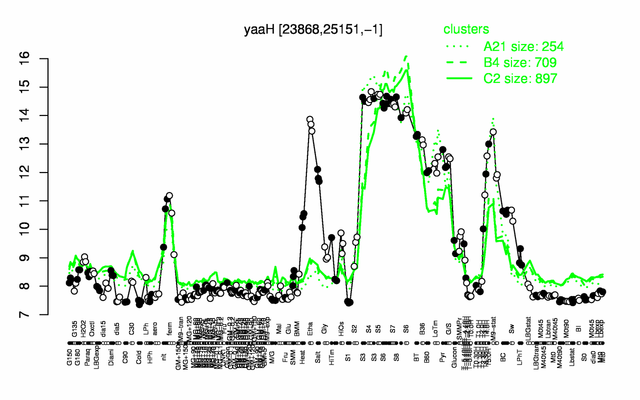
| |
Contents
Categories containing this gene/protein
sporulation proteins, general stress proteins (controlled by SigB)
This gene is a member of the following regulons
SigB regulon, SigE regulon, SpoIIID regulon
The gene
Basic information
- Locus tag: BSU00160
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- contains a N-acetylglucosamine-polymer-binding LysM domain PubMed
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P37531
- KEGG entry: [3]
- E.C. number:
Additional information
required for survival of ethanol stress PubMed
Expression and regulation
- Operon: yaaH PubMed
- Regulation:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications