Difference between revisions of "PksL"
| Line 37: | Line 37: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 65: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| Line 83: | Line 77: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 109: | Line 103: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[pksC]]-[[pksD]]-[[pksE]]-[[acpK]]-[[pksF]]-[[pksG]]-[[pksH]]-[[pksI]]-[[pksJ]]-[[pksL]]-[[pksM]]-[[pksN]]-[[pksR]]'' {{PubMed|22383849}} |
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pksL_1807921_1821537_1 pksL] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=pksL_1807921_1821537_1 pksL] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** expressed during the transition from growth to stationary phase ([[AbrB]], [[CodY]]) {{PubMed|24187085}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[AbrB]]: transcription repression {{PubMed|24187085}} | ||
| + | ** [[CodY]]: transcription activation {{PubMed|24187085}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 140: | Line 137: | ||
=References= | =References= | ||
| − | + | <pubmed> 22383849 24187085</pubmed> | |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:48, 18 December 2013
- Description: polyketide synthase of type I
| Gene name | pksL |
| Synonyms | pksX, pksA, outG |
| Essential | no |
| Product | polyketide synthase of type I |
| Function | polyketide synthesis |
| Gene expression levels in SubtiExpress: pksL | |
| MW, pI | 505 kDa, 5.197 |
| Gene length, protein length | 13614 bp, 4538 aa |
| Immediate neighbours | pksJ, pksM |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed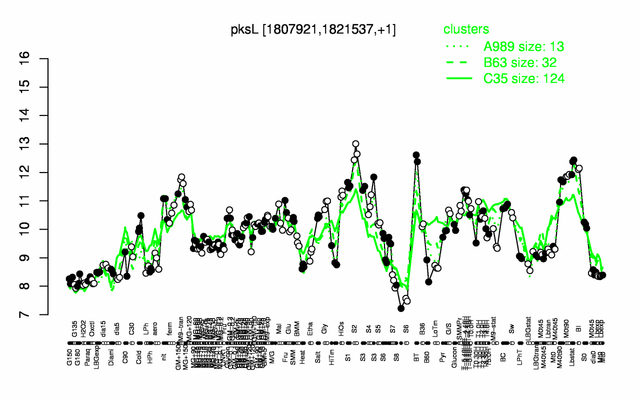
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU17190
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: Q05470
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References