Difference between revisions of "Hag"
(→Database entries) |
|||
| Line 103: | Line 103: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId= 3A5X 3A5X] (from ''Salmonella typhimurium'', 42% identity) {{PubMed|23804755}} | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=3A5X 3A5X] (from ''Salmonella typhimurium'', 42% identity) {{PubMed|23804755}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P02968 P02968] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P02968 P02968] | ||
Revision as of 16:09, 29 July 2013
- Description: flagellin protein, about 20,000 subunits make up one flagellum
| Gene name | hag |
| Synonyms | |
| Essential | no |
| Product | flagellin protein |
| Function | motility and chemotaxis |
| Gene expression levels in SubtiExpress: hag | |
| Interactions involving this protein in SubtInteract: Hag | |
| MW, pI | 32 kDa, 4.782 |
| Gene length, protein length | 912 bp, 304 aa |
| Immediate neighbours | yvyC, csrA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 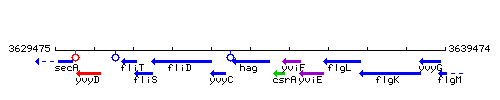
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed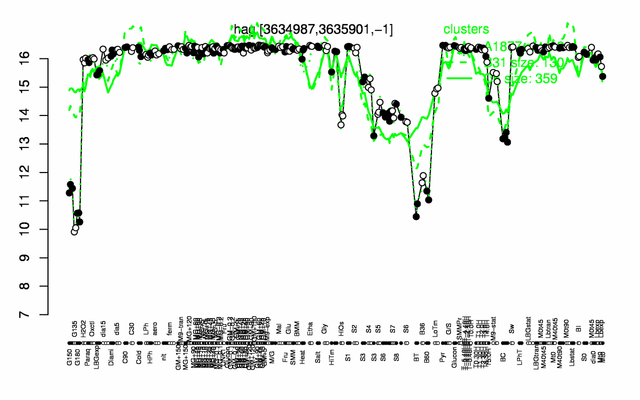
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
CodY regulon, CsrA regulon, ScoC regulon, SigD regulon
The gene
Basic information
- Locus tag: BSU35360
Phenotypes of a mutant
No swarming motility on B medium. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: bacterial flagellin family (according to Swiss-Prot)
- Paralogous protein(s): YvzB (C-terminal domain of Hag)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P02968
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP901 (aphA3), GP902 (tet), both available in Stülke lab
- lacZ fusion: pGP1035 (in pAC6), pGP755 (in pAC7), there is also a series of promoter deletion variants in pAC6 and pAC7, all available in Stülke lab
- GFP fusion: BP494 (bglS:: (hag-promoter-cfp-aphA3)), BP496 (amyE:: (hag-promoter-iyfp-cat)), available in Stülke lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Sampriti Mukherjee, Paul Babitzke, Daniel B Kearns
FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis.
J Bacteriol: 2013, 195(2);297-306
[PubMed:23144244]
[WorldCat.org]
[DOI]
(I p)
Sampriti Mukherjee, Helen Yakhnin, Dave Kysela, Josh Sokoloski, Paul Babitzke, Daniel B Kearns
CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis.
Mol Microbiol: 2011, 82(2);447-61
[PubMed:21895793]
[WorldCat.org]
[DOI]
(I p)
Christine Diethmaier, Nico Pietack, Katrin Gunka, Christoph Wrede, Martin Lehnik-Habrink, Christina Herzberg, Sebastian Hübner, Jörg Stülke
A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation.
J Bacteriol: 2011, 193(21);5997-6007
[PubMed:21856853]
[WorldCat.org]
[DOI]
(I p)
Kassem Hamze, Sabine Autret, Krzysztof Hinc, Soumaya Laalami, Daria Julkowska, Romain Briandet, Margareth Renault, Cédric Absalon, I Barry Holland, Harald Putzer, Simone J Séror
Single-cell analysis in situ in a Bacillus subtilis swarming community identifies distinct spatially separated subpopulations differentially expressing hag (flagellin), including specialized swarmers.
Microbiology (Reading): 2011, 157(Pt 9);2456-2469
[PubMed:21602220]
[WorldCat.org]
[DOI]
(I p)
Gert Bange, Nico Kümmerer, Christoph Engel, Gunes Bozkurt, Klemens Wild, Irmgard Sinning
FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system.
Proc Natl Acad Sci U S A: 2010, 107(25);11295-300
[PubMed:20534509]
[WorldCat.org]
[DOI]
(I p)
Kassem Hamze, Daria Julkowska, Sabine Autret, Krzysztof Hinc, Krzysztofa Nagorska, Agnieszka Sekowska, I Barry Holland, Simone J Séror
Identification of genes required for different stages of dendritic swarming in Bacillus subtilis, with a novel role for phrC.
Microbiology (Reading): 2009, 155(Pt 2);398-412
[PubMed:19202088]
[WorldCat.org]
[DOI]
(P p)
Prashant Kodgire, K Krishnamurthy Rao
hag expression in Bacillus subtilis is both negatively and positively regulated by ScoC.
Microbiology (Reading): 2009, 155(Pt 1);142-149
[PubMed:19118355]
[WorldCat.org]
[DOI]
(P p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Helen Yakhnin, Pallavi Pandit, Tom J Petty, Carol S Baker, Tony Romeo, Paul Babitzke
CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding.
Mol Microbiol: 2007, 64(6);1605-20
[PubMed:17555441]
[WorldCat.org]
[DOI]
(P p)
D B Mirel, W F Estacio, M Mathieu, E Olmsted, J Ramirez, L M Márquez-Magaña
Environmental regulation of Bacillus subtilis sigma(D)-dependent gene expression.
J Bacteriol: 2000, 182(11);3055-62
[PubMed:10809682]
[WorldCat.org]
[DOI]
(P p)
T Caramori, A Galizzi
The UP element of the promoter for the flagellin gene, hag, stimulates transcription from both SigD- and SigA-dependent promoters in Bacillus subtilis.
Mol Gen Genet: 1998, 258(4);385-8
[PubMed:9648743]
[WorldCat.org]
[DOI]
(P p)
Y F Chen, J D Helmann
DNA-melting at the Bacillus subtilis flagellin promoter nucleates near -10 and expands unidirectionally.
J Mol Biol: 1997, 267(1);47-59
[PubMed:9096206]
[WorldCat.org]
[DOI]
(P p)
K Fredrick, T Caramori, Y F Chen, A Galizzi, J D Helmann
Promoter architecture in the flagellar regulon of Bacillus subtilis: high-level expression of flagellin by the sigma D RNA polymerase requires an upstream promoter element.
Proc Natl Acad Sci U S A: 1995, 92(7);2582-6
[PubMed:7708689]
[WorldCat.org]
[DOI]
(P p)
D B Mirel, M J Chamberlin
The Bacillus subtilis flagellin gene (hag) is transcribed by the sigma 28 form of RNA polymerase.
J Bacteriol: 1989, 171(6);3095-101
[PubMed:2498284]
[WorldCat.org]
[DOI]
(P p)