Difference between revisions of "YtxM"
| Line 24: | Line 24: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[menB]]'', ''[[menD]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[menB]]'', ''[[menD]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU30810 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU30810 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU30810 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU30810 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU30810 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU30810 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:ytxM_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:ytxM_context.gif]] | ||
Revision as of 11:05, 14 May 2013
- Description: similar to prolyl aminopeptidase
| Gene name | ytxM |
| Synonyms | ytfB |
| Essential | no |
| Product | unknown |
| Function | ubiquinone and menaquinone biosynthesis |
| Gene expression levels in SubtiExpress: ytxM | |
| Metabolic function and regulation of this protein in SubtiPathways: Menaquinone | |
| MW, pI | 30 kDa, 7.175 |
| Gene length, protein length | 822 bp, 274 aa |
| Immediate neighbours | menB, menD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 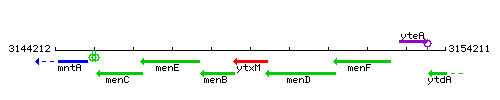
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed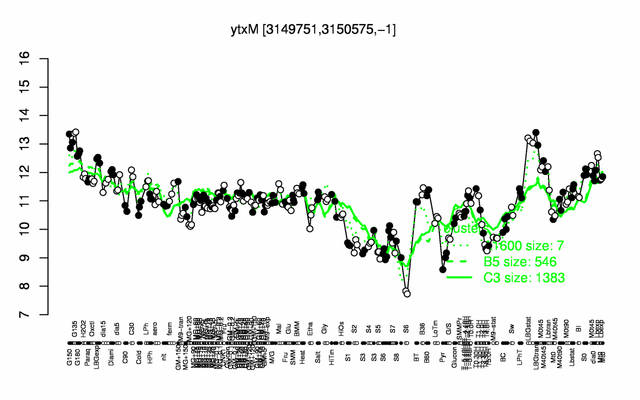
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU30810
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: lipase/esterase LIP3/bchO family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P23974
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
M M Mehanni, A P Turnbull, S E Sedelnikova, P J Baker, S Foster, D W Rice
Crystallization and preliminary X-ray analysis of the ytxM gene product from Bacillus subtilis.
Acta Crystallogr D Biol Crystallogr: 2002, 58(Pt 12);2138-40
[PubMed:12454479]
[WorldCat.org]
[DOI]
(P p)
H Antelmann, C Scharf, M Hecker
Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis.
J Bacteriol: 2000, 182(16);4478-90
[PubMed:10913081]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)
X Qin, H W Taber
Transcriptional regulation of the Bacillus subtilis menp1 promoter.
J Bacteriol: 1996, 178(3);705-13
[PubMed:8550504]
[WorldCat.org]
[DOI]
(P p)
J R Driscoll, H W Taber
Sequence organization and regulation of the Bacillus subtilis menBE operon.
J Bacteriol: 1992, 174(15);5063-71
[PubMed:1629163]
[WorldCat.org]
[DOI]
(P p)