Difference between revisions of "Sandbox"
Raphael2215 (talk | contribs) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' enolase, glycolytic/ gluconeogenic enzyme, [[universally conserved protein]]<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''eno'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | + | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || enolase |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || enzyme in glycolysis/ gluconeogenesis |
| − | |||
|- | |- | ||
| − | |style="background:# | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/Eno Eno] |
|- | |- | ||
| − | |style="background:# | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|''' | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 46,4 kDa, 4.49 |
|- | |- | ||
| − | | | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 1290 bp, 430 amino acids |
|- | |- | ||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yvbK]]'', ''[[pgm]]'' |
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB15395]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' | ||
| + | |- | ||
| + | |colspan="2" | '''Genetic context''' <br/> [[Image:eno_context.gif]] | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
| − | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=eno_3476555_3477847_-1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:eno_expression.png|500px]] | |
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id= | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | <br/><br/><br/><br/> | ||
__TOC__ | __TOC__ | ||
| Line 38: | Line 42: | ||
| − | <br/><br/> | + | |
| + | |||
| + | <br/><br/><br/><br/> | ||
| + | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[carbon core metabolism]]}}, |
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[membrane proteins]]}}, |
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| − | {{SubtiWiki category|[[ | + | {{SubtiWiki category|[[universally conserved proteins]]}} |
| − | |||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| − | {{SubtiWiki regulon|[[ | + | {{SubtiWiki regulon|[[CggR regulon]]}} |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=The gene= | =The gene= | ||
=== Basic information === | === Basic information === | ||
| − | + | * '''Locus tag:''' BSU33900 | |
| − | * '''Locus tag:''' | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| + | * no growth on LB, requires glucose and malate | ||
| + | * essential according to Kobayashi et al. on LB [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed] | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/cggR-gapA-pgk-tpiA-pgm-eno.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10899] |
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 78: | Line 77: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' 2-phospho-D-glycerate = phosphoenolpyruvate + H<sub>2</sub>O (according to Swiss-Prot) 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O |
| − | * '''Protein family:''' | + | * '''Protein family:''' enolase family (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 86: | Line 85: | ||
=== Extended information on the protein === | === Extended information on the protein === | ||
| − | * '''Kinetic information:''' | + | * '''Kinetic information:''' reversible Michaelis-Menten [http://www.ncbi.nlm.nih.gov/sites/entrez/25885 PubMed] |
* '''Domains:''' | * '''Domains:''' | ||
| + | ** substrate binding domain (366–369) | ||
| − | * '''Modification:''' | + | * '''Modification:''' phosphorylation on Thr-141 AND Ser-259 AND Tyr-281 AND Ser-325 [http://www.ncbi.nlm.nih.gov/sites/entrez/17218307 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed] |
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' Mg2+ |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| + | ** Inhibited by EDTA [http://www.ncbi.nlm.nih.gov/sites/entrez/25885 PubMed] | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** forms octamers {{PubMed|22198292}} | ||
| + | ** part of the [[RNA degradosome]] {{PubMed|19193632}} | ||
| + | ** [[Eno]]-[[PfkA]] {{PubMed|19193632}}, K(D) of the interaction: 40 nM {{PubMed|22198292}} | ||
| + | ** [[Eno]]-[[Rny]] {{PubMed|19193632,21803996}}, K(D) of the interaction: 100 nM {{PubMed|22198292}} | ||
| + | ** [[Eno]]-[[CshA]] {{PubMed|20572937}} | ||
| − | * '''[[Localization]]:''' | + | * '''[[Localization]]:''' |
| − | ** membrane associated [http://www.ncbi.nlm.nih.gov/ | + | ** cytoplasm [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] |
| + | ** membrane associated [http://www.ncbi.nlm.nih.gov/sites/entrez/18763711 PubMed] | ||
| + | ** exported, this requires a long, unbent α-helix (from A108 to L126) {{PubMed|15003462,21856851}} | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' |
| + | ** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=4A3R 4A3R] {{PubMed|22198292}} | ||
| + | ** [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1W6T 1W6T] (from ''Streptococcus pneumoniae'') {{PubMed|15476816}} | ||
| − | * '''UniProt:''' [http://www.uniprot.org/uniprot/ | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/P37869 P37869] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu: | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU33900] |
| − | * '''E.C. number:''' | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/4.2.1.11 4.2.1.11] |
=== Additional information=== | === Additional information=== | ||
| + | * Enolase is a [[moonlighting proteins|moonlighting protein]]. [http://www.ncbi.nlm.nih.gov/sites/entrez/19193632 PubMed] | ||
| + | * There are indications that this enzyme is an octamer [http://www.ncbi.nlm.nih.gov/sites/entrez/25885 PubMed] | ||
| + | * [[universally conserved protein]] | ||
| + | * extensive information on the structure and enzymatic properties of Eno can be found at [http://www.proteopedia.org/wiki/index.php/Enolase Proteopedia] | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | |||
| − | * ''' | + | * '''Operon:''' |
| + | ** ''[[cggR]]-[[gapA]]-[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ||
| + | ** ''[[pgk]]-[[tpiA]]-[[pgm]]-[[eno]]'' {{PubMed|11489127}} | ||
| − | * ''' | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=eno_3476555_3477847_-1 eno] {{PubMed|22383849}} |
| + | |||
| + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=eno_3476555_3477847_-1 eno] {{PubMed|22383849}} | ||
| + | |||
| + | * '''Sigma factor:''' [[SigA]] {{PubMed|11489127}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| − | ** | + | ** expression activated by glucose (3.3 fold) [http://www.ncbi.nlm.nih.gov/pubmed/12850135 PubMed] |
| − | ** | + | ** ''[[cggR]]'': induced by glycolytic substrates [[CggR]] {{PubMed|11489127}} |
| − | ** | + | ** ''[[pgk]]'': constitutive {{PubMed|11489127}} |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' transcription repression by [[CggR]] {{PubMed|11489127}} |
| − | |||
| − | |||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 133: | Line 150: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' |
| + | ** GP594 (''eno''::''cat''), available in [[Stülke]] lab | ||
| + | ** GP599 (''eno''::''erm''), available in [[Stülke]] lab | ||
| + | ** GP698 (''eno''-''[[pgm]]''::''cat''), available in [[Stülke]] lab | ||
| + | |||
| + | * '''Expression vector:''' | ||
| + | ** pGP1426 (expression of ''[[eno]]'' in ''B. subtilis'', in [[pBQ200]]), available in [[Stülke]] lab | ||
| + | ** pGP1500 (expression of ''[[pgm]]'' and ''[[eno]]'' in ''B. subtilis'', in [[pBQ200]]), available in [[Stülke]] lab | ||
| + | ** pGP563 (N-terminal His-tag, in [[pWH844]]), available in [[Stülke]] lab | ||
| + | ** pGP1276 (N-terminal Strep-tag, purification from ''E. coli'', in [[pGP172]]), available in [[Stülke]] lab | ||
| + | ** pGP93 (N-terminal Strep-tag, purification from ''B. subtilis'', for [[SPINE]], in [[pGP380]]), available in [[Stülke]] lab | ||
| + | ** GP1215 (''eno''-''Strep'' ''(spc)''), purification from ''B. subtilis'', for [[SPINE]], available in [[Stülke]] lab | ||
| − | |||
| − | |||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| + | ** see ''[[pgk]]'' | ||
| + | |||
| + | * '''GFP fusion:''' pHT315-yfp-eno, available in [[Mijakovic]] lab | ||
| − | * ''' | + | * '''two-hybrid system:''' ''B. pertussis'' adenylate cyclase-based bacterial two hybrid system ([[BACTH]]), available in [[Stülke]] lab |
| − | * ''' | + | * '''FLAG-tag construct:''' GP1214 (spc, based on [[pGP1331]]), available in the [[Stülke]] lab |
| − | * '''Antibody:''' | + | * '''Antibody:''' available in [[Stülke]] lab |
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Stülke|Jörg Stülke]], University of Göttingen, Germany | ||
| + | [http://wwwuser.gwdg.de/~genmibio/stuelke.html Homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
=References= | =References= | ||
| − | '''Additional publications:''' {{PubMed| | + | ==Reviews== |
| − | <pubmed> | + | <pubmed> 8994873 </pubmed> |
| + | ==Subcellular localization of enolase== | ||
| + | '''Additional publications:''' {{PubMed|21856851}} | ||
| + | <pubmed> 16479537 18763711 15003462 20497499 </pubmed> | ||
| + | |||
| + | ==Other original publications== | ||
| + | <pubmed> 17726680, 17218307, 12850135, 19193632, 11489127, 8021172, 17505547, 25885, 20572937 15476816 9988532 ,21803996 22198292 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 11:39, 18 April 2012
- Description: enolase, glycolytic/ gluconeogenic enzyme, universally conserved protein
| Gene name | eno | ||
| Synonyms | |||
| Essential | no | ||
| Product | enolase | ||
| Function | enzyme in glycolysis/ gluconeogenesis | ||
| Interactions involving this protein in SubtInteract: Eno | |||
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |||
| MW, pI | 46,4 kDa, 4.49 | ||
| Gene length, protein length | 1290 bp, 430 amino acids | ||
| Immediate neighbours | yvbK, pgm | ||
| Get the DNA and protein sequences (Barbe et al., 2009) | |||
Genetic context 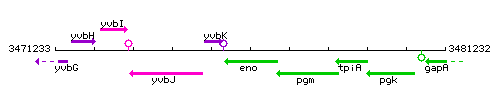
This image was kindly provided by SubtiList
|
Expression at a glance PubMed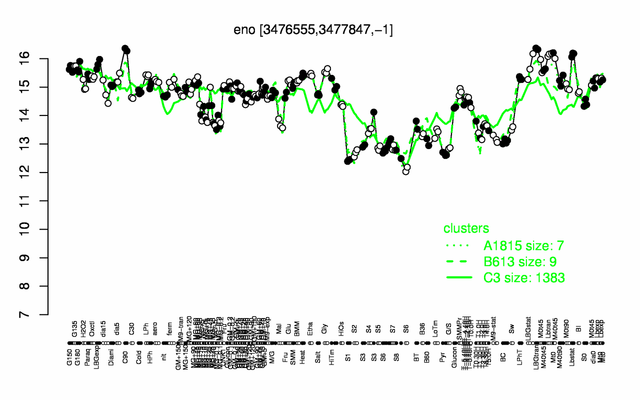
| ||
Contents
Categories containing this gene/protein
carbon core metabolism, membrane proteins, phosphoproteins, universally conserved proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33900
Phenotypes of a mutant
- no growth on LB, requires glucose and malate
- essential according to Kobayashi et al. on LB PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H2O (according to Swiss-Prot) 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: reversible Michaelis-Menten PubMed
- Domains:
- substrate binding domain (366–369)
- Cofactor(s): Mg2+
- Effectors of protein activity:
- Inhibited by EDTA PubMed
Database entries
- UniProt: P37869
- KEGG entry: [3]
- E.C. number: 4.2.1.11
Additional information
- Enolase is a moonlighting protein. PubMed
- There are indications that this enzyme is an octamer PubMed
- universally conserved protein
- extensive information on the structure and enzymatic properties of Eno can be found at Proteopedia
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- pGP1426 (expression of eno in B. subtilis, in pBQ200), available in Stülke lab
- pGP1500 (expression of pgm and eno in B. subtilis, in pBQ200), available in Stülke lab
- pGP563 (N-terminal His-tag, in pWH844), available in Stülke lab
- pGP1276 (N-terminal Strep-tag, purification from E. coli, in pGP172), available in Stülke lab
- pGP93 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Stülke lab
- GP1215 (eno-Strep (spc)), purification from B. subtilis, for SPINE, available in Stülke lab
- lacZ fusion:
- see pgk
- GFP fusion: pHT315-yfp-eno, available in Mijakovic lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody: available in Stülke lab
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Subcellular localization of enolase
Additional publications: PubMed
Carsten Jers, Malene Mejer Pedersen, Dafni Katerina Paspaliari, Wolfgang Schütz, Christina Johnsson, Boumediene Soufi, Boris Macek, Peter Ruhdal Jensen, Ivan Mijakovic
Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates.
Mol Microbiol: 2010, 77(2);287-99
[PubMed:20497499]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Grégory Boël, Vianney Pichereau, Ivan Mijakovic, Alain Mazé, Sandrine Poncet, Sylvie Gillet, Jean-Christophe Giard, Axel Hartke, Yanick Auffray, Josef Deutscher
Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export?
J Mol Biol: 2004, 337(2);485-96
[PubMed:15003462]
[WorldCat.org]
[DOI]
(P p)
Other original publications