Difference between revisions of "TagU"
| Line 40: | Line 40: | ||
{{SubtiWiki regulon|[[LytR regulon]]}}, | {{SubtiWiki regulon|[[LytR regulon]]}}, | ||
{{SubtiWiki regulon|[[SigX regulon]]}} | {{SubtiWiki regulon|[[SigX regulon]]}} | ||
| − | + | =The [[LytR regulon]]:= | |
| + | * ''[[lytA]]-[[lytB]]-[[lytC]]'', ''[[lytR]]'' | ||
=The gene= | =The gene= | ||
Revision as of 11:25, 18 December 2010
| Gene name | lytR |
| Synonyms | |
| Essential | no |
| Product | negative regulator for lytA-lytB-lytC and lytR expression |
| Function | regulation of cell separation and autolysis |
| MW, pI | 34 kDa, 9.527 |
| Gene length, protein length | 918 bp, 306 aa |
| Immediate neighbours | lytA, mnaA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 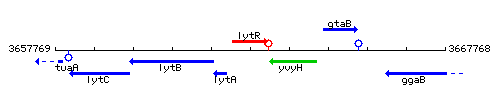
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
cell wall degradation/ turnover, transcription factors and their control, cell envelope stress proteins (controlled by SigM, W, X, Y), membrane proteins
This gene is a member of the following regulons
The LytR regulon:
The gene
Basic information
- Locus tag: BSU35650
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: lytR/cpsA/psr regulatory protein family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: Q02115
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: lytR PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
X Huang, J D Helmann
Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus-directed search.
J Mol Biol: 1998, 279(1);165-73
[PubMed:9636707]
[WorldCat.org]
[DOI]
(P p)
V Lazarevic, P Margot, B Soldo, D Karamata
Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier.
J Gen Microbiol: 1992, 138(9);1949-61
[PubMed:1357079]
[WorldCat.org]
[DOI]
(P p)