Difference between revisions of "DacF"
(→Expression and regulation) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' penicillin-binding protein I, D-alanyl-D-alanine carboxypeptidase <br/><br/> | + | * '''Description:''' [[penicillin-binding protein]] I, D-alanyl-D-alanine carboxypeptidase <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || penicillin-binding protein I, <br/>D-alanyl-D-alanine carboxypeptidase | + | |style="background:#ABCDEF;" align="center"| '''Product''' || [[penicillin-binding protein]] I, <br/>D-alanyl-D-alanine carboxypeptidase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || carboxypeptidase | |style="background:#ABCDEF;" align="center"|'''Function''' || carboxypeptidase | ||
Revision as of 13:44, 18 November 2009
- Description: penicillin-binding protein I, D-alanyl-D-alanine carboxypeptidase
| Gene name | dacF |
| Synonyms | |
| Essential | no |
| Product | penicillin-binding protein I, D-alanyl-D-alanine carboxypeptidase |
| Function | carboxypeptidase |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 43 kDa, 9.912 |
| Gene length, protein length | 1167 bp, 389 aa |
| Immediate neighbours | spoIIAA, pupG |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 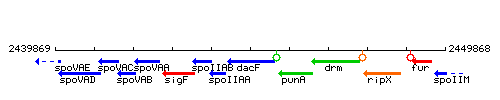
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU23480
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Preferential cleavage: (Ac)(2)-L-Lys-D-Ala-|-D-Ala (according to Swiss-Prot)
- Protein family: peptidase S11 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P38422
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
D L Popham, M E Gilmore, P Setlow
Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties.
J Bacteriol: 1999, 181(1);126-32
[PubMed:9864321]
[WorldCat.org]
[DOI]
(P p)
Peter J Lewis, Jeffery Errington
Use of green fluorescent protein for detection of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis.
Microbiology (Reading): 1996, 142 ( Pt 4);733-740
[PubMed:8936302]
[WorldCat.org]
[DOI]
(P p)