Difference between revisions of "SpoIIR"
(→References) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' required for processing of pro-SigE <br/><br/> | + | * '''Description:''' required for processing of pro-[[SigE]] <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || morphological chekcpoint for SigE processing | + | |style="background:#ABCDEF;" align="center"| '''Product''' || morphological chekcpoint for [[SigE]] processing |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || control of SigE activation | + | |style="background:#ABCDEF;" align="center"|'''Function''' || control of [[SigE]] activation |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 25 kDa, 4.737 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 25 kDa, 4.737 | ||
Revision as of 11:31, 28 July 2009
- Description: required for processing of pro-SigE
| Gene name | spoIIR |
| Synonyms | ipc-27d, csfX |
| Essential | no |
| Product | morphological chekcpoint for SigE processing |
| Function | control of SigE activation |
| MW, pI | 25 kDa, 4.737 |
| Gene length, protein length | 672 bp, 224 aa |
| Immediate neighbours | ywlB, ywlA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 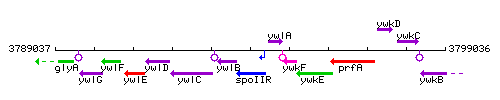
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU36970
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: P39151
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Avigdor Eldar, Vasant K Chary, Panagiotis Xenopoulos, Michelle E Fontes, Oliver C Losón, Jonathan Dworkin, Patrick J Piggot, Michael B Elowitz
Partial penetrance facilitates developmental evolution in bacteria.
Nature: 2009, 460(7254);510-4
[PubMed:19578359]
[WorldCat.org]
[DOI]
(I p)
Stephanie T Wang, Barbara Setlow, Erin M Conlon, Jessica L Lyon, Daisuke Imamura, Tsutomu Sato, Peter Setlow, Richard Losick, Patrick Eichenberger
The forespore line of gene expression in Bacillus subtilis.
J Mol Biol: 2006, 358(1);16-37
[PubMed:16497325]
[WorldCat.org]
[DOI]
(P p)
Aileen Rubio, Kit Pogliano
Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis.
EMBO J: 2004, 23(7);1636-46
[PubMed:15044948]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, Richard Losick
An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis.
Mol Microbiol: 2002, 43(1);27-38
[PubMed:11849534]
[WorldCat.org]
[DOI]
(P p)
A E Hofmeister, A Londoño-Vallejo, E Harry, P Stragier, R Losick
Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis.
Cell: 1995, 83(2);219-26
[PubMed:7585939]
[WorldCat.org]
[DOI]
(P p)