Difference between revisions of "ClpC"
(→Database entries) |
|||
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation | |style="background:#ABCDEF;" align="center"|'''Function''' || protein degradation | ||
| − | positive regulator of autolysin (LytC and LytD) synthesis | + | positive regulator of autolysin ([[LytC]] and [[LytD]]) synthesis |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 89 kDa, 5.746 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 89 kDa, 5.746 | ||
| Line 71: | Line 71: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[YpbH]]-[[ClpC]], [[McsB]]-[[ClpC]], [[ClpC]]-[[MecA]], [[ClpC]]-[[ClpP]] | + | * '''Interactions:''' [[YpbH]]-[[ClpC]], [[McsB]]-[[ClpC]], [[ClpC]]-[[MecA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/10447896 PubMed], [[ClpC]]-[[ClpP]] |
* '''Localization:''' cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heatshock, colocalization with [[ClpP]] [http://www.ncbi.nlm.nih.gov/pubmed/18786145 Pubmed] | * '''Localization:''' cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heatshock, colocalization with [[ClpP]] [http://www.ncbi.nlm.nih.gov/pubmed/18786145 Pubmed] | ||
Revision as of 14:19, 23 May 2009
- Description: ATP-dependent Clp protease, ATPase subunit
| Gene name | clpC |
| Synonyms | mecB |
| Essential | no |
| Product | ATP-dependent Clp protease, ATPase subunit |
| Function | protein degradation |
| MW, pI | 89 kDa, 5.746 |
| Gene length, protein length | 2430 bp, 810 aa |
| Immediate neighbours | mcsB, radA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 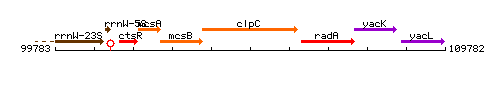
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATPase/chaperone
- Protein family: mecA family (according to Swiss-Prot) clpA/clpB family. ClpC subfamily (according to Swiss-Prot), AAA+ -type ATPase (IPR013093) InterPro (PF07724) PFAM
Extended information on the protein
- Kinetic information:
- Domains: AAA-ATPase PFAM
- Modification:
- Cofactor(s):
- Effectors of protein activity:
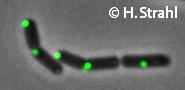
- Localization: cytoplasmic polar clusters, excluded from the nucleoid, induced clustering upon heatshock, colocalization with ClpP Pubmed
Database entries
- Structure: 2K77 (N-terminal domain)
- Swiss prot entry: P37571
- KEGG entry: BSU00860
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: clpC::tet available from the Hamoen] Lab
- Expression vector:
- lacZ fusion:
- GFP fusion: C-terminal GFP fusions (single copy, also as CFP and YFP variants) available from the Hamoen] Lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Newcastle University, UK homepage
Your additional remarks
References
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed