Difference between revisions of "AraD"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' L-ribulose-phosphate 4-epimerase <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || L-ribulose | + | |style="background:#ABCDEF;" align="center"| '''Product''' || L-ribulose-phosphate 4-epimerase |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || arabinose utilization | |style="background:#ABCDEF;" align="center"|'''Function''' || arabinose utilization | ||
Revision as of 17:59, 18 March 2009
- Description: L-ribulose-phosphate 4-epimerase
| Gene name | araD |
| Synonyms | |
| Essential | no |
| Product | L-ribulose-phosphate 4-epimerase |
| Function | arabinose utilization |
| MW, pI | 25 kDa, 5.243 |
| Gene length, protein length | 687 bp, 229 aa |
| Immediate neighbours | araL, araB |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 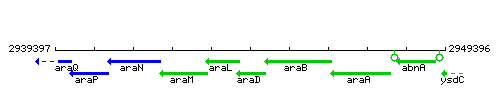
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry:
- E.C. number:
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Inácio JM, Costa C, de Sá-Nogueira I. (2003) Distinct molecular mechanisms involved in carbon catabolite repression of the arabinose regulon in Bacillus subtilis. Microbiology. Sep;149(Pt 9): 2345-55. PubMed
- Sa-Nogueira I.M.G., Nogueira T.V., Soares S., de Lencastre H. (1997) The Bacillus subtilis L-arabinose (ara) operon: nucleotide sequence, genetic organization and expression. Microbiology. Mar;143(Pt 3): 957-69. PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed