Difference between revisions of "FliG"
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || flagellar | + | |style="background:#ABCDEF;" align="center"| '''Product''' || flagellar C ring protein |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || movement and chemotaxis | |style="background:#ABCDEF;" align="center"|'''Function''' || movement and chemotaxis | ||
Revision as of 12:19, 4 March 2015
- Description: flagellar basal-body protein, membrane anchor of the basal body, flagellar motor switch protein, physically transduces force from MotA to the rotation of FliF
| Gene name | fliG |
| Synonyms | flaA15 |
| Essential | no |
| Product | flagellar C ring protein |
| Function | movement and chemotaxis |
| Gene expression levels in SubtiExpress: fliG | |
| Interactions involving this protein in SubtInteract: FliG | |
| MW, pI | 38 kDa, 4.46 |
| Gene length, protein length | 1014 bp, 338 aa |
| Immediate neighbours | fliF, fliH |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 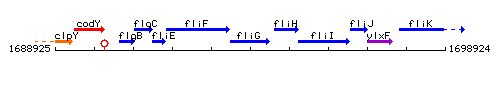
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed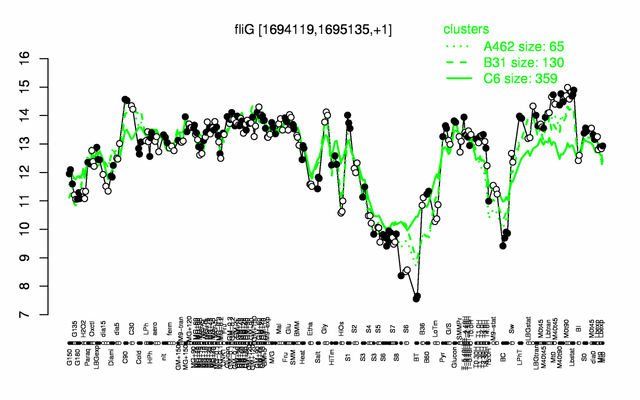
| |
Contents
Categories containing this gene/protein
motility and chemotaxis, membrane proteins
This gene is a member of the following regulons
CodY regulon, DegU regulon, SigD regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU16220
Phenotypes of a mutant
- mucoid phenotype due to the overproduction of poly-gamma-glutamate PubMed
- no secretion of FlgM, permanent inhibition of SigD PubMed
Database entries
- BsubCyc: BSU16220
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: fliG family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity: interaction with EpsE prevents energetization of the flagellum by MotA-MotB PubMed
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU16220
- Structure:
- UniProt: P23448
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: part of the fla-che operon (see there for regulation)
- Regulation: see fla-che operon
- Regulatory mechanism: see fla-che operon
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 250 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References