Difference between revisions of "RecR"
| Line 1: | Line 1: | ||
* '''Description:''' required for the formation of [[RecA]] DNA repair centers | * '''Description:''' required for the formation of [[RecA]] DNA repair centers | ||
<br/><br/> | <br/><br/> | ||
| − | |||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
Revision as of 11:19, 4 June 2014
- Description: required for the formation of RecA DNA repair centers
| Gene name | recR |
| Synonyms | recM |
| Essential | no |
| Product | formation of RecA DNA repair centers |
| Function | DNA repair/ recombination |
| Gene expression levels in SubtiExpress: recR | |
| Interactions involving this protein in SubtInteract: RecR | |
| MW, pI | 21 kDa, 5.301 |
| Gene length, protein length | 594 bp, 198 aa |
| Immediate neighbours | yaaK, yaaL |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 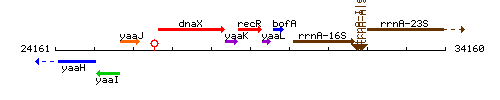
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed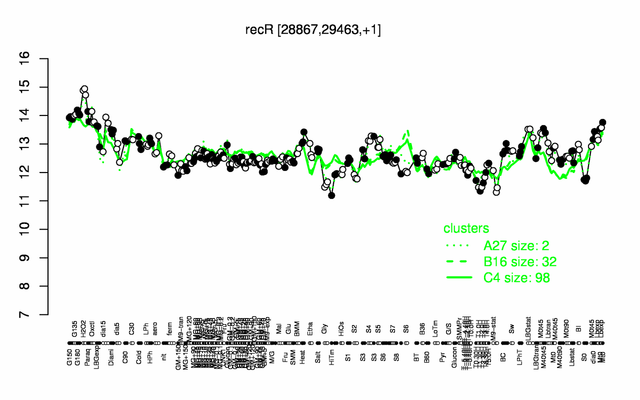
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00210
Phenotypes of a mutant
- drastically reduced survival of mature dormant spores after exposure to ultrahigh vacuum desiccation and ionizing radiation that induce single strand (ss) DNA nicks and double-strand breaks (DSBs) PubMed
Database entries
- BsubCyc: BSU00210
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: recR family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (homogeneous) PubMed
Database entries
- BsubCyc: BSU00210
- Structure:
- UniProt: P24277
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information: the mRNA is very stable (half-life > 15 min) PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Original publications
Justin S Lenhart, Eileen R Brandes, Jeremy W Schroeder, Roderick J Sorenson, Hollis D Showalter, Lyle A Simmons
RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress.
J Bacteriol: 2014, 196(15);2851-60
[PubMed:24891441]
[WorldCat.org]
[DOI]
(I p)
Ignacija Vlašić, Ramona Mertens, Elena M Seco, Begoña Carrasco, Silvia Ayora, Günther Reitz, Fabian M Commichau, Juan C Alonso, Ralf Moeller
Bacillus subtilis RecA and its accessory factors, RecF, RecO, RecR and RecX, are required for spore resistance to DNA double-strand break.
Nucleic Acids Res: 2014, 42(4);2295-307
[PubMed:24285298]
[WorldCat.org]
[DOI]
(I p)
Joanna Timmins, Ingar Leiros, Sean McSweeney
Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding.
EMBO J: 2007, 26(13);3260-71
[PubMed:17581636]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)