Difference between revisions of "Tkt"
| Line 45: | Line 45: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[carbon core metabolism]]}}, | {{SubtiWiki category|[[carbon core metabolism]]}}, | ||
| − | {{SubtiWiki category|[[phosphoproteins]]}} | + | {{SubtiWiki category|[[phosphoproteins]]}}, |
| + | [[most abundant proteins]] | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| Line 65: | Line 66: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
| − | |||
| − | |||
=The protein= | =The protein= | ||
| Line 83: | Line 81: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylated on ser/ thr/ tyr [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed] | * '''Modification:''' phosphorylated on ser/ thr/ tyr [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed] | ||
| Line 122: | Line 120: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** belongs to the 100 [[most abundant proteins]] {{PubMed|15378759}} | ||
=Biological materials = | =Biological materials = | ||
| Line 149: | Line 148: | ||
<pubmed>9924800 </pubmed> | <pubmed>9924800 </pubmed> | ||
==Original publications== | ==Original publications== | ||
| − | <pubmed>14651647,16493705, 19603213 9068642 16143417 15375635 19193632 23568212 23965678 </pubmed> | + | <pubmed>14651647,16493705, 19603213 9068642 16143417 15375635 19193632 23568212 23965678 15378759</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:43, 5 March 2014
- Description: transketolase
| Gene name | tkt |
| Synonyms | tktA |
| Essential | no |
| Product | transketolase |
| Function | pentose phosphate pathway |
| Gene expression levels in SubtiExpress: tkt | |
| Interactions involving this protein in SubtInteract: Tkt | |
| Metabolic function and regulation of this protein in SubtiPathways: tkt | |
| MW, pI | 72 kDa, 4.803 |
| Gene length, protein length | 2001 bp, 667 aa |
| Immediate neighbours | ynzC, sirA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 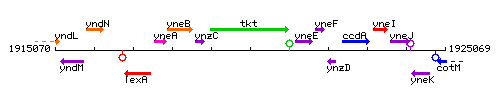
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed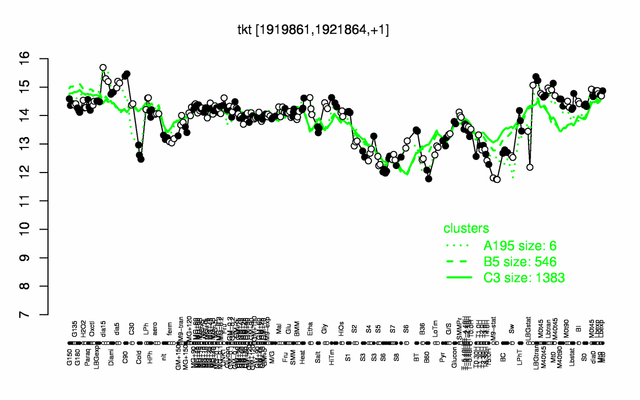
| |
Contents
Categories containing this gene/protein
carbon core metabolism, phosphoproteins, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU17890
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = D-ribose 5-phosphate + D-xylulose 5-phosphate (according to Swiss-Prot)
- Protein family: transketolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylated on ser/ thr/ tyr PubMed
- Cofactors: thiamine pyrophosphate
- Effectors of protein activity:
Database entries
- Structure: [3K95], complex with thiamine diphosphate, from B. anthracis
- UniProt: P45694
- KEGG entry: [3]
- E.C. number: 2.2.1.1
Additional information
Expression and regulation
- Operon: tkt PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
Biological materials
- Mutant: BS4530 (aphA3), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Stefan Lüdtke, Piotr Neumann, Karl M Erixon, Finian Leeper, Ronald Kluger, Ralf Ficner, Kai Tittmann
Sub-ångström-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem: 2013, 5(9);762-7
[PubMed:23965678]
[WorldCat.org]
[DOI]
(I p)
Yong-Cheol Park, Hae-Jin Lee, Chang Sup Kim, Jin-Ho Seo
Effects of oxygen supply and mixed sugar concentration on D-ribose production by a transketolase-deficient Bacillus subtilis SPK1.
J Microbiol Biotechnol: 2013, 23(4);560-4
[PubMed:23568212]
[WorldCat.org]
[DOI]
(I p)
Lin Wu, Zhimin Li, Qin Ye
Enhanced D-ribose biosynthesis in batch culture of a transketolase-deficient Bacillus subtilis strain by citrate.
J Ind Microbiol Biotechnol: 2009, 36(10);1289-96
[PubMed:19603213]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Fabian M Rothe, Christina Herzberg, Eva Wagner, Daniel Hellwig, Martin Lehnik-Habrink, Elke Hammer, Uwe Völker, Jörg Stülke
Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing.
Mol Cell Proteomics: 2009, 8(6);1350-60
[PubMed:19193632]
[WorldCat.org]
[DOI]
(I p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
Yong-Cheol Park, Jin-Ho Choi, George N Bennett, Jin-Ho Seo
Characterization of D-ribose biosynthesis in Bacillus subtilis JY200 deficient in transketolase gene.
J Biotechnol: 2006, 121(4);508-16
[PubMed:16143417]
[WorldCat.org]
[DOI]
(P p)
Christine Eymann, Annette Dreisbach, Dirk Albrecht, Jörg Bernhardt, Dörte Becher, Sandy Gentner, Le Thi Tam, Knut Büttner, Gerrit Buurman, Christian Scharf, Simone Venz, Uwe Völker, Michael Hecker
A comprehensive proteome map of growing Bacillus subtilis cells.
Proteomics: 2004, 4(10);2849-76
[PubMed:15378759]
[WorldCat.org]
[DOI]
(P p)
Yong-Cheol Park, Sung-Gun Kim, Kyungmoon Park, Kelvin H Lee, Jin-Ho Seo
Fed-batch production of D-ribose from sugar mixtures by transketolase-deficient Bacillus subtilis SPK1.
Appl Microbiol Biotechnol: 2004, 66(3);297-302
[PubMed:15375635]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
T Schiött, C von Wachenfeldt, L Hederstedt
Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis.
J Bacteriol: 1997, 179(6);1962-73
[PubMed:9068642]
[WorldCat.org]
[DOI]
(P p)