Difference between revisions of "SpoIVFA"
| Line 72: | Line 72: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' PDZ protease {{PubMed|24243021}} |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| Line 81: | Line 81: | ||
* '''Domains:''' | * '''Domains:''' | ||
| + | ** the ectodomain of [[SpoIVFA]] is cleaved off by [[SpoIVB]] (first cleavage) {{PubMed|24243021}} | ||
| + | ** the domains that inhibits [[SpoIVFB]] is cleaved off by [[CtpB]] (second cleavage) {{PubMed|24243021}} | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 91: | Line 93: | ||
** [[SpoIVFB]]-[[SpoIVFA]], to facilitate the recruitment of [[BofA]] to [[SpoIVFB]] {{PubMed|11959848}} | ** [[SpoIVFB]]-[[SpoIVFA]], to facilitate the recruitment of [[BofA]] to [[SpoIVFB]] {{PubMed|11959848}} | ||
** [[BofA]]-[[SpoIVFA]] {{PubMed|11959848}} | ** [[BofA]]-[[SpoIVFA]] {{PubMed|11959848}} | ||
| + | ** [[SpoIVB]]-[[SpoIVFA]] {{PubMed|24243021}} | ||
| + | ** [[CtpB]]-[[SpoIVFA]] {{PubMed|24243021}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| − | ** | + | ** mother cell membrane {{PubMed|24243021}} |
** integral membrane protein {{PubMed|11959848}} | ** integral membrane protein {{PubMed|11959848}} | ||
| Line 107: | Line 111: | ||
=== Additional information=== | === Additional information=== | ||
| − | + | ** the ectodomain of [[SpoIVFA]] is cleaved off by [[SpoIVB]] (first cleavage) {{PubMed|24243021}} | |
| − | cleaved by [[SpoIVB]] | + | ** the domains that inhibits [[SpoIVFB]] is cleaved off by [[CtpB]] (second cleavage) {{PubMed|24243021} |
=Expression and regulation= | =Expression and regulation= | ||
| Line 124: | Line 128: | ||
** [[SpoIIID]]: transcription repression {{PubMed|15383836}} | ** [[SpoIIID]]: transcription repression {{PubMed|15383836}} | ||
| − | * '''Additional information:''' cleaved by [[SpoIVB]] | + | * '''Additional information:''' |
| + | ** the ectodomain of [[SpoIVFA]] is cleaved off by [[SpoIVB]] (first cleavage) {{PubMed|24243021}} | ||
| + | ** the domains that inhibits [[SpoIVFB]] is cleaved off by [[CtpB]] (second cleavage) {{PubMed|24243021}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 10:03, 3 December 2013
- Description: inhibitor of SpoIVFB metalloprotease
| Gene name | spoIVFA |
| Synonyms | bofB, spoVL |
| Essential | no |
| Product | inhibitor of SpoIVFB metalloprotease |
| Function | control of SigK activation |
| Gene expression levels in SubtiExpress: spoIVFA | |
| Interactions involving this protein in SubtInteract: SpoIVFA | |
| MW, pI | 29 kDa, 7.292 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | spoIVFB, minD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 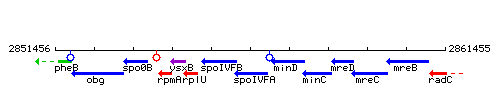
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed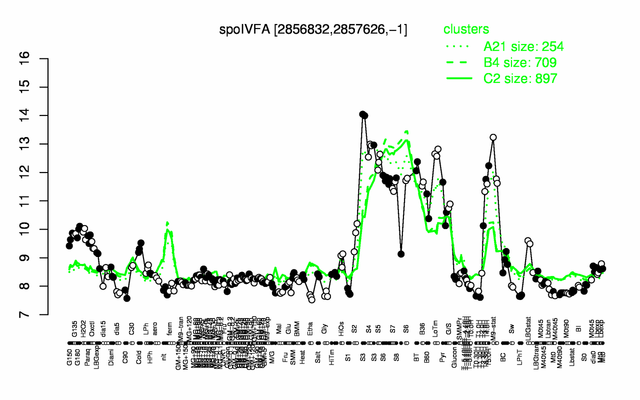
| |
Contents
Categories containing this gene/protein
sigma factors and their control, sporulation proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU27980
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: PDZ protease PubMed
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P26936
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Nathalie Campo, David Z Rudner
SpoIVB and CtpB are both forespore signals in the activation of the sporulation transcription factor sigmaK in Bacillus subtilis.
J Bacteriol: 2007, 189(16);6021-7
[PubMed:17557826]
[WorldCat.org]
[DOI]
(P p)
Nathalie Campo, David Z Rudner
A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis.
Mol Cell: 2006, 23(1);25-35
[PubMed:16818230]
[WorldCat.org]
[DOI]
(P p)
Thierry Doan, Kathleen A Marquis, David Z Rudner
Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum.
Mol Microbiol: 2005, 55(6);1767-81
[PubMed:15752199]
[WorldCat.org]
[DOI]
(P p)
Leif Steil, Mónica Serrano, Adriano O Henriques, Uwe Völker
Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis.
Microbiology (Reading): 2005, 151(Pt 2);399-420
[PubMed:15699190]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
Tran C Dong, Simon M Cutting
The PDZ domain of the SpoIVB transmembrane signaling protein enables cis-trans interactions involving multiple partners leading to the activation of the pro-sigmaK processing complex in Bacillus subtilis.
J Biol Chem: 2004, 279(42);43468-78
[PubMed:15292188]
[WorldCat.org]
[DOI]
(P p)
Tran C Dong, Simon M Cutting
SpoIVB-mediated cleavage of SpoIVFA could provide the intercellular signal to activate processing of Pro-sigmaK in Bacillus subtilis.
Mol Microbiol: 2003, 49(5);1425-34
[PubMed:12940997]
[WorldCat.org]
[DOI]
(P p)
David Z Rudner, Richard Losick
A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization.
Genes Dev: 2002, 16(8);1007-18
[PubMed:11959848]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, R Losick
Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1998, 95(6);3162-7
[PubMed:9501233]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, S Alper, R Losick
Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis.
Genes Cells: 1996, 1(6);529-42
[PubMed:9078383]
[WorldCat.org]
[DOI]
(P p)
E Ricca, S Cutting, R Losick
Characterization of bofA, a gene involved in intercompartmental regulation of pro-sigma K processing during sporulation in Bacillus subtilis.
J Bacteriol: 1992, 174(10);3177-84
[PubMed:1577688]
[WorldCat.org]
[DOI]
(P p)
S Cutting, S Roels, R Losick
Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis.
J Mol Biol: 1991, 221(4);1237-56
[PubMed:1942049]
[WorldCat.org]
[DOI]
(P p)
S Cutting, V Oke, A Driks, R Losick, S Lu, L Kroos
A forespore checkpoint for mother cell gene expression during development in B. subtilis.
Cell: 1990, 62(2);239-50
[PubMed:2115401]
[WorldCat.org]
[DOI]
(P p)