Difference between revisions of "YrhJ"
(→References) |
|||
| Line 147: | Line 147: | ||
=References= | =References= | ||
| − | + | <pubmed>11734890,10917605 ,11574077,14741768,15122913, 12775685 16716428 16381045 20525796 12207695 9636707 21926231</pubmed> | |
| − | <pubmed>11734890,10917605 ,11574077,14741768,15122913, 12775685 16716428 16381045 20525796 12207695 9636707</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:04, 23 October 2013
- Description: cytochrome P450 (CYP102A3)/ NADPH-cytochrome P450 reductase
| Gene name | yrhJ |
| Synonyms | cypE |
| Essential | no |
| Product | NADPH-cytochrome P450 reductase |
| Function | fatty acid metabolism |
| Gene expression levels in SubtiExpress: yrhJ | |
| MW, pI | 118 kDa, 6.017 |
| Gene length, protein length | 3162 bp, 1054 aa |
| Immediate neighbours | rrcM, fatR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 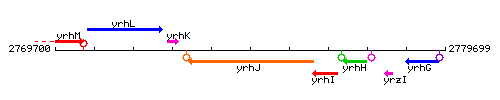
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
electron transport/ other, lipid metabolism/ other, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
FatR regulon, SigM regulon, SigW regulon, SigX regulon
The gene
Basic information
- Locus tag: BSU27160
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- hydroxylates medium-chain fatty acids in subterminal positions PubMed
- Protein family:
- Paralogous protein(s): YetO
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
Database entries
- Structure: 2X7Y (from Bacillus megaterium; 67% identity, 88% similarity)
- UniProt: O08336
- KEGG entry: [3]
- E.C. number: 1.6.2.4
Additional information
Expression and regulation
- Regulation:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References