Difference between revisions of "WprA"
| Line 23: | Line 23: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yisL]]'', ''[[yisN]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yisL]]'', ''[[yisN]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU10770 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU10770 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU10770 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU10770 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU10770 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU10770 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:wprA_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:wprA_context.gif]] | ||
Revision as of 09:52, 14 May 2013
- Description: secreted quality control protease
| Gene name | wprA |
| Synonyms | yisM |
| Essential | no |
| Product | secreted quality control protease |
| Function | protein quality control |
| Gene expression levels in SubtiExpress: wprA | |
| Interactions involving this protein in SubtInteract: WprA | |
| MW, pI | 96 kDa, 9.58 |
| Gene length, protein length | 2682 bp, 894 aa |
| Immediate neighbours | yisL, yisN |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 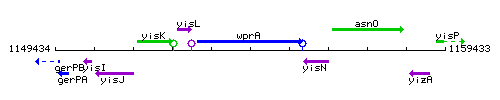
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall/ other, proteolysis,
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10770
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: peptidase S8 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P54423
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- An antisense RNA is predicted forwprA PubMed
- the amount of the mRNA is substantially decreased upon depletion of RNase Y PubMed
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Carmine G Monteferrante, Calum MacKichan, Elodie Marchadier, Maria-Victoria Prejean, Rut Carballido-López, Jan Maarten van Dijl
Mapping the twin-arginine protein translocation network of Bacillus subtilis.
Proteomics: 2013, 13(5);800-11
[PubMed:23180473]
[WorldCat.org]
[DOI]
(I p)
Laxmi Krishnappa, Carmine G Monteferrante, Jan Maarten van Dijl
Degradation of the twin-arginine translocation substrate YwbN by extracytoplasmic proteases of Bacillus subtilis.
Appl Environ Microbiol: 2012, 78(21);7801-4
[PubMed:22923395]
[WorldCat.org]
[DOI]
(I p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Lehnik-Habrink M, Schaffer M, Mäder U, Diethmaier C, Herzberg C, Stülke J RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol. 2011 81(6): 1459-1473. PubMed:21815947