Difference between revisions of "Eno"
Raphael2215 (talk | contribs) |
|||
| Line 26: | Line 26: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yvbK]]'', ''[[pgm]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yvbK]]'', ''[[pgm]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU33900 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU33900 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU33900 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU33900 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU33900 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU33900 DNA_with_flanks] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:eno_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:eno_context.gif]] | ||
Revision as of 11:17, 14 May 2013
- Description: enolase, glycolytic/ gluconeogenic enzyme, universally conserved protein
| Gene name | eno |
| Synonyms | |
| Essential | Yes (PubMed) |
| Product | enolase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| Gene expression levels in SubtiExpress: eno | |
| Interactions involving this protein in SubtInteract: Eno | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 46,4 kDa, 4.49 |
| Gene length, protein length | 1290 bp, 430 amino acids |
| Immediate neighbours | yvbK, pgm |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 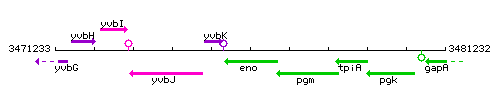
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, essential genes, membrane proteins, phosphoproteins, universally conserved proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33900
Phenotypes of a mutant
- no growth on LB, requires glucose and malate
- essential according to Kobayashi et al. on LB PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = phosphoenolpyruvate + H2O (according to Swiss-Prot) 2-phospho-D-glycerate = phosphoenolpyruvate + H(2)O
- Protein family: enolase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: reversible Michaelis-Menten PubMed
- Domains:
- substrate binding domain (366–369)
- Cofactor(s): Mg2+
- Effectors of protein activity:
- Inhibited by EDTA PubMed
Database entries
- UniProt: P37869
- KEGG entry: [3]
- E.C. number: 4.2.1.11
Additional information
- Enolase is a moonlighting protein. PubMed
- There are indications that this enzyme is an octamer PubMed
- universally conserved protein
- extensive information on the structure and enzymatic properties of Eno can be found at Proteopedia
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- GP594 (eno::cat), available in Jörg Stülke's lab
- GP599 (eno::erm), available in Jörg Stülke's lab
- GP698 (eno-pgm::cat), available in Jörg Stülke's lab
- Expression vector:
- pGP1426 (expression of eno in B. subtilis, in pBQ200), available in Jörg Stülke's lab
- pGP1500 (expression of pgm and eno in B. subtilis, in pBQ200), available in Jörg Stülke's lab
- pGP563 (N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- pGP1276 (N-terminal Strep-tag, purification from E. coli, in pGP172), available in Jörg Stülke's lab
- pGP93 (N-terminal Strep-tag, purification from B. subtilis, for SPINE, in pGP380), available in Jörg Stülke's lab
- GP1215 (chromosomal eno-Strep fusion, spc), purification from B. subtilis, for SPINE, available in Jörg Stülke's lab
- lacZ fusion:
- see pgk
- GFP fusion:
- pHT315-yfp-eno, available in Mijakovic lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- FLAG-tag construct:
- GP1214 (spc, based on pGP1331), available in Jörg Stülke's lab
- Antibody: available in Jörg Stülke's lab
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Reviews
Subcellular localization of enolase
Additional publications: PubMed
Carsten Jers, Malene Mejer Pedersen, Dafni Katerina Paspaliari, Wolfgang Schütz, Christina Johnsson, Boumediene Soufi, Boris Macek, Peter Ruhdal Jensen, Ivan Mijakovic
Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates.
Mol Microbiol: 2010, 77(2);287-99
[PubMed:20497499]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Other original publications