Difference between revisions of "ManA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 99: | Line 99: | ||
* '''Operon:''' ''[[manP]]-[[manA]]-[[yjdF]]'' {{PubMed|20139185}} | * '''Operon:''' ''[[manP]]-[[manA]]-[[yjdF]]'' {{PubMed|20139185}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=manA_1274692_1275639_1 manA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigA]] {{PubMed|20139185}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 108: | Line 110: | ||
** [[ManR]]: transcription activation in the presence of mannose and absence of glucose {{PubMed|20139185}} | ** [[ManR]]: transcription activation in the presence of mannose and absence of glucose {{PubMed|20139185}} | ||
| − | * '''Additional information:''' | + | * '''Additional information:''' |
=Biological materials = | =Biological materials = | ||
Revision as of 18:05, 12 April 2012
- Description: mannose-6-phosphate isomerase, required for proper cell wall synthesis
| Gene name | manA |
| Synonyms | yjdE |
| Essential | no |
| Product | mannose-6-phosphate isomerase |
| Function | mannose utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 35 kDa, 5.279 |
| Gene length, protein length | 945 bp, 315 aa |
| Immediate neighbours | manP, yjdF |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 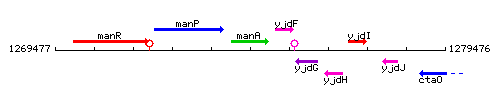
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU12020
Phenotypes of a mutant
- cells lose the characteristic rod shape typical of wild type B. subtilis cells and instead appear as elongated spheres, which are significantly larger than normal PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31646
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Maya Elbaz, Sigal Ben-Yehuda
The metabolic enzyme ManA reveals a link between cell wall integrity and chromosome morphology.
PLoS Genet: 2010, 6(9);e1001119
[PubMed:20862359]
[WorldCat.org]
[DOI]
(I e)
Tianqi Sun, Josef Altenbuchner
Characterization of a mannose utilization system in Bacillus subtilis.
J Bacteriol: 2010, 192(8);2128-39
[PubMed:20139185]
[WorldCat.org]
[DOI]
(I p)
M S Turner, J D Helmann
Mutations in multidrug efflux homologs, sugar isomerases, and antimicrobial biosynthesis genes differentially elevate activity of the sigma(X) and sigma(W) factors in Bacillus subtilis.
J Bacteriol: 2000, 182(18);5202-10
[PubMed:10960106]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)