Difference between revisions of "Pdp"
(→Extended information on the protein) |
|||
| Line 81: | Line 81: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' | + | * '''[[SubtInteract|Interactions]]:''' |
| − | * '''Localization:''' | + | * '''[[Localization]]:''' |
=== Database entries === | === Database entries === | ||
Revision as of 11:58, 8 November 2011
- Description: pyrimidine nucleoside phosphorylase
| Gene name | pdp |
| Synonyms | |
| Essential | no |
| Product | pyrimidine nucleoside phosphorylase |
| Function | pyrimidine interconversion |
| Metabolic function and regulation of this protein in SubtiPathways: Nucleoside catabolism, Nucleotides (regulation) | |
| MW, pI | 46 kDa, 4.765 |
| Gene length, protein length | 1299 bp, 433 aa |
| Immediate neighbours | hutM, nupC |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 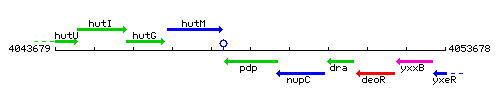
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39400
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: A pyrimidine nucleoside + phosphate = a pyrimidine base + alpha-D-ribose 1-phosphate (according to Swiss-Prot)
- Protein family: thymidine/pyrimidine-nucleoside phosphorylase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1BRW (Geobacillus stearothermophilus)
- UniProt: P39142
- KEGG entry: [3]
- E.C. number: 2.4.2.2
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Xianmin Zeng, Anne Galinier, Hans H Saxild
Catabolite repression of dra-nupC-pdp operon expression in Bacillus subtilis.
Microbiology (Reading): 2000, 146 ( Pt 11);2901-2908
[PubMed:11065368]
[WorldCat.org]
[DOI]
(P p)
Y Miwa, A Nakata, A Ogiwara, M Yamamoto, Y Fujita
Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis.
Nucleic Acids Res: 2000, 28(5);1206-10
[PubMed:10666464]
[WorldCat.org]
[DOI]
(I p)
X Zeng, H H Saxild
Identification and characterization of a DeoR-specific operator sequence essential for induction of dra-nupC-pdp operon expression in Bacillus subtilis.
J Bacteriol: 1999, 181(6);1719-27
[PubMed:10074062]
[WorldCat.org]
[DOI]
(P p)
H H Saxild, L N Andersen, K Hammer
Dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein.
J Bacteriol: 1996, 178(2);424-34
[PubMed:8550462]
[WorldCat.org]
[DOI]
(P p)