Difference between revisions of "GmuB"
| Line 50: | Line 50: | ||
| + | |||
| + | = Categories containing this gene/protein = | ||
| + | {{SubtiWiki category|[[phosphotransferase systems]]}}, | ||
| + | {{SubtiWiki category|[[utilization of specific carbon sources]]}}, | ||
| + | {{SubtiWiki category|[[phosphoproteins]]}} | ||
=The protein= | =The protein= | ||
Revision as of 17:07, 30 November 2010
- Description: glucomannan-specific phosphotransferase system, EIIB component of the PTS
| Gene name | gmuB |
| Synonyms | ydhM |
| Essential | no |
| Product | glucomannan-specific phosphotransferase system EIIB component |
| Function | glucomannan uptake and phosphorylation |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 11 kDa, 4.724 |
| Gene length, protein length | 309 bp, 103 aa |
| Immediate neighbours | pbuE, gmuA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 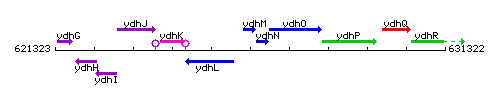
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU05810
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources, phosphoproteins
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- UniProt: O05505
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Yoshito Sadaie, Hisashi Nakadate, Reiko Fukui, Lii Mien Yee, Kei Asai
Glucomannan utilization operon of Bacillus subtilis.
FEMS Microbiol Lett: 2008, 279(1);103-9
[PubMed:18177310]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)