Difference between revisions of "GudB"
(→Biological materials) |
|||
| Line 80: | Line 80: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3K8Z 3K8Z] (enzymatically active GudB1) | + | * '''Structure:''' [http://www.rcsb.org/pdb/search/structidSearch.do?structureId=3K8Z 3K8Z] (enzymatically active GudB1) {{PubMed|20630473}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P50735 P50735] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P50735 P50735] | ||
| Line 131: | Line 131: | ||
=References= | =References= | ||
| − | <pubmed>18603778,9829940 ,17183217 18723616, 18326565 </pubmed> | + | <pubmed>18603778,9829940 ,17183217 18723616, 18326565 20630473</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:32, 16 July 2010
- Description: trigger enzyme: glutamate dehydrogenase (cryptic in 168 and derivatives)
| Gene name | gudB |
| Synonyms | ypcA |
| Essential | no |
| Product | trigger enzyme: glutamate dehydrogenase |
| Function | glutamate utilization, control of GltC activity |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 47 kDa, 5.582 |
| Gene length, protein length | 1278 bp, 426 aa |
| Immediate neighbours | ypdA, ypbH |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 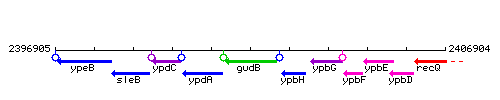
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU22960
Phenotypes of a mutant
The gene is cryptic. If gudB is activated (gudB1 mutation), the bacteria are able to utilize glutamate as the only carbon source. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamate + H2O + NAD+ = 2-oxoglutarate + NH3 + NADH (according to Swiss-Prot)
- Protein family: Glu/Leu/Phe/Val dehydrogenases family (according to Swiss-Prot)
- Paralogous protein(s): RocG
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: P50735
- KEGG entry: [3]
- E.C. number: 1.4.1.2
Additional information
Expression and regulation
- Operon: gudB PubMed
- Regulation: constitutively expressed
- Regulatory mechanism:
- Additional information: GudB is subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: GP691 (cat), available in Stülke lab
- Expression vector:
- FLAG-tag construct: GP1194 (gudB, spc, based on pGP1331), GP1195 (gudB1, spc, based on pGP1331), available in Stülke lab
- GFP fusion:
- two-hybrid system:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
The GudB protein is active in other legacy B. subtilis strains (e.g. strain 122). Thus, it can be speculated that the ancestral gudB gene was not cryptic, but became so as a product of the "domestication" of B. subtilis 168 in the lab. PubMed
References