Difference between revisions of "PucC"
(→Database entries) |
|||
| Line 78: | Line 78: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1JRO 1JRO] (from ''Rhodobacter capsulatus'', 39% identity, 55% similarity) {{PubMed|11796116}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O32145 O32145] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O32145 O32145] | ||
Revision as of 12:17, 19 February 2010
- Description: xanthine dehydrogenase
| Gene name | pucC |
| Synonyms | yurD |
| Essential | no |
| Product | xanthine dehydrogenase |
| Function | purine utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Purine salvage, Purine catabolism, Nucleotides (regulation) | |
| MW, pI | 29 kDa, 8.576 |
| Gene length, protein length | 831 bp, 277 aa |
| Immediate neighbours | pucD, pucB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 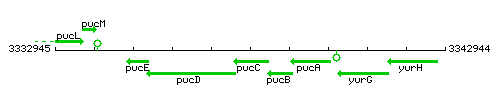
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU32490
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Xanthine + NAD+ + H2O = urate + NADH (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: O32145
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ken-ichi Yoshida, Hirotake Yamaguchi, Masaki Kinehara, Yo-hei Ohki, Yoshiko Nakaura, Yasutaro Fujita
Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box.
Mol Microbiol: 2003, 49(1);157-65
[PubMed:12823818]
[WorldCat.org]
[DOI]
(P p)
Lars Beier, Per Nygaard, Hanne Jarmer, Hans H Saxild
Transcription analysis of the Bacillus subtilis PucR regulon and identification of a cis-acting sequence required for PucR-regulated expression of genes involved in purine catabolism.
J Bacteriol: 2002, 184(12);3232-41
[PubMed:12029039]
[WorldCat.org]
[DOI]
(P p)
A C Schultz, P Nygaard, H H Saxild
Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator.
J Bacteriol: 2001, 183(11);3293-302
[PubMed:11344136]
[WorldCat.org]
[DOI]
(P p)