Difference between revisions of "Pgm"
| Line 36: | Line 36: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU33910 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 82: | Line 82: | ||
* '''Swiss prot entry:''' [http://www.expasy.ch/cgi-bin/sprot-search-ac?P39773] | * '''Swiss prot entry:''' [http://www.expasy.ch/cgi-bin/sprot-search-ac?P39773] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU33910] |
* '''E.C. number:''' [http://www.expasy.org/enzyme/5.4.2.1 5.4.2.1] 5.4.2.1] | * '''E.C. number:''' [http://www.expasy.org/enzyme/5.4.2.1 5.4.2.1] 5.4.2.1] | ||
Revision as of 11:55, 3 June 2009
- Description: phosphoglycerate mutase, glycolytic / gluconeogenic enzyme
| Gene name | pgm |
| Synonyms | gpmI |
| Essential | yes |
| Product | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
| Function | enzyme in glycolysis / gluconeogenesis |
| MW, pI | 56,1 kDa, 5.21 |
| Gene length, protein length | 1533 bp, 511 amino acids |
| Immediate neighbours | tpi, eno |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 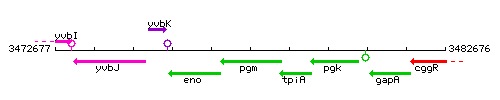
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU33910
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = 3-phospho-D-glycerate (according to Swiss-Prot) 2-phospho-D-glycerate = 3-phospho-D-glycerate
- Protein family: BPG-independent phosphoglycerate mutase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Cofactor(s): 2 manganese ions per subunit
- Effectors of protein activity: inhibited by heavy-metal ions, 2,3-butanedione and sulfhydryl agents PubMed
- Interactions: Pgm-PfkA
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure: 1EJJ (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), 1EQJ (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), Geobacillus stearothermophilus, complex with 2-phosphoglycerate NCBI, Geobacillus stearothermophilus, complex with 3-phosphoglycerate NCBI
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: 5.4.2.1 5.4.2.1]
Additional information
is pH sensitive
Expression and regulation
- Sigma factor: SigA
- Regulation: expression activated by glucose (7.3 fold) PubMed
cggR: neg. regulated by CggR PubMed, induced by sugar
- Additional information:
Biological materials
- Mutant:
- Expression vector: pGP1101 (N-terminal His-tag, in pWH844), pGP396 (Pgm-S62A, N-terminal His-tag, in pWH844), pGP92 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Stülke lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Mark J. Jedrzejas, Research Center Oakland, CA, USA Homepage
Your additional remarks
References
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Masatoshi Nukui, Luciane V Mello, James E Littlejohn, Barbara Setlow, Peter Setlow, Kijeong Kim, Terrance Leighton, Mark J Jedrzejas
Structure and molecular mechanism of Bacillus anthracis cofactor-independent phosphoglycerate mutase: a crucial enzyme for spores and growing cells of Bacillus species.
Biophys J: 2007, 92(3);977-88
[PubMed:17085493]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Daniel J Rigden, Ejvis Lamani, Luciane V Mello, James E Littlejohn, Mark J Jedrzejas
Insights into the catalytic mechanism of cofactor-independent phosphoglycerate mutase from X-ray crystallography, simulated dynamics and molecular modeling.
J Mol Biol: 2003, 328(4);909-20
[PubMed:12729763]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, P Setlow
Comparison of the binuclear metalloenzymes diphosphoglycerate-independent phosphoglycerate mutase and alkaline phosphatase: their mechanism of catalysis via a phosphoserine intermediate.
Chem Rev: 2001, 101(3);607-18
[PubMed:11712498]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, M Chander, P Setlow, G Krishnasamy
Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J Biol Chem: 2000, 275(30);23146-53
[PubMed:10764795]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, M Chander, P Setlow, G Krishnasamy
Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J: 2000, 19(7);1419-31
[PubMed:10747010]
[WorldCat.org]
[DOI]
(P p)
M Chander, P Setlow, E Lamani, M J Jedrzejas
Structural studies on a 2,3-diphosphoglycerate independent phosphoglycerate mutase from Bacillus stearothermophilus.
J Struct Biol: 1999, 126(2);156-65
[PubMed:10388626]
[WorldCat.org]
[DOI]
(P p)
M A Leyva-Vazquez, P Setlow
Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis.
J Bacteriol: 1994, 176(13);3903-10
[PubMed:8021172]
[WorldCat.org]
[DOI]
(P p)