Difference between revisions of "AbrB"
(New page: * '''Description:''' write here <br/><br/> {| align="right" border="1" cellpadding="2" |- |style="background:#ABCDEF;" align="center"|'''Gene name''' |''abrB'' |- |style="background:#ABC...) |
|||
| Line 12: | Line 12: | ||
|style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional regulator | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional regulator | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of transition state genes | + | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of transition state genes |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 10 kDa, 6.57 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 10 kDa, 6.57 | ||
| Line 56: | Line 56: | ||
* '''Protein family:''' | * '''Protein family:''' | ||
| − | * '''Paralogous protein(s):''' | + | * '''Paralogous protein(s):''' [[Abh]] |
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 68: | Line 68: | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' | + | * '''Effectors of protein activity:''' interaction with [[AbbA]] results in inactivation of AbrB [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed] |
| − | * '''Interactions:''' | + | * '''Interactions:''' [[AbrB]]-[[AbbA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed] |
* '''Localization:''' | * '''Localization:''' | ||
| Line 85: | Line 85: | ||
=== Additional information=== | === Additional information=== | ||
| + | |||
| + | negative regulation of ''[[abrB]], [[aprE]], [[ftsAZ]], [[kinC]], [[motA]], [[nprE]], [[pbpE]], [[rbs]], [[spo0H]], [[spoVG]], [[spo0E]], [[tycA]], [[sbo]]-[[alb]], [[yqxM]]-[[sipW]]-[[tasA]]''; | ||
| + | positive regulation of ''[[comK]], [[hpr]]'' | ||
| + | |||
=Expression and regulation= | =Expression and regulation= | ||
| Line 92: | Line 96: | ||
* '''Sigma factor:''' | * '''Sigma factor:''' | ||
| − | * '''Regulation:''' | + | * '''Regulation:''' expressed at the onset of staionary phase |
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' repressed by [[Spo0A]]-P |
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 100: | Line 104: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' TT731 (aphA3) |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 113: | Line 117: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | [[Richard Losick]], Harvard Univ., Cambridge, USA | ||
| + | [[Mark Strauch]], | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 118: | Line 125: | ||
=References= | =References= | ||
| + | # Banse et al. (2008) Parallel pathways of repression and antirepression governing the transition to stationary phase in ''Bacillus subtilis''.''Proc. Natl. Acad. Sci. USA'' '''105:''' 15547-15552. [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 11:27, 1 February 2009
- Description: write here
| Gene name | abrB |
| Synonyms | cpsX |
| Essential | |
| Product | transcriptional regulator |
| Function | regulation of transition state genes |
| MW, pI | 10 kDa, 6.57 |
| Gene length, protein length | 288 bp, 96 aa |
| Immediate neighbours | |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 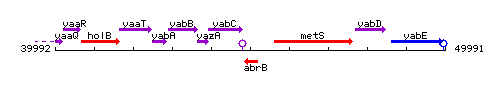
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): Abh
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry:
- E.C. number:
Additional information
negative regulation of abrB, aprE, ftsAZ, kinC, motA, nprE, pbpE, rbs, spo0H, spoVG, spo0E, tycA, sbo-alb, yqxM-sipW-tasA; positive regulation of comK, hpr
Expression and regulation
- Operon:
- Sigma factor:
- Regulation: expressed at the onset of staionary phase
- Regulatory mechanism: repressed by Spo0A-P
- Additional information:
Biological materials
- Mutant: TT731 (aphA3)
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Richard Losick, Harvard Univ., Cambridge, USA Mark Strauch,