Difference between revisions of "MtlA"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' trigger enzyme: mannitol-specific phosphotransferase system, EIICBA <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || trigger enzyme: mannitol-specific phosphotransferase system, EIICBA |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || mannitol uptake | + | |style="background:#ABCDEF;" align="center"|'''Function''' || mannitol uptake and phosphorylation, control of MtlR activity |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 65 kDa, 8.778 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 65 kDa, 8.778 | ||
Revision as of 16:54, 16 March 2009
- Description: trigger enzyme: mannitol-specific phosphotransferase system, EIICBA
| Gene name | mtlA |
| Synonyms | |
| Essential | no |
| Product | trigger enzyme: mannitol-specific phosphotransferase system, EIICBA |
| Function | mannitol uptake and phosphorylation, control of MtlR activity |
| MW, pI | 65 kDa, 8.778 |
| Gene length, protein length | 1830 bp, 610 aa |
| Immediate neighbours | ycnL, mtlD |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 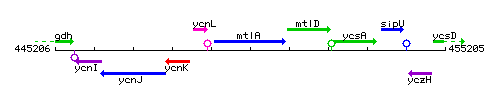
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation on Ser-559 PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry:
- E.C. number:
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: