Difference between revisions of "PhoP"
| Line 1: | Line 1: | ||
| − | * '''Description:''' [[two-component systems|two-component]] response regulator, regulation of phosphate metabolism <br/><br/> | + | * '''Description:''' [[two-component systems|two-component]] response regulator ([[transcription factors of the OmpR family|OmpR family]]), regulation of phosphate metabolism <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || [[two-component systems|two-component]] response regulator | + | |style="background:#ABCDEF;" align="center"| '''Product''' || [[two-component systems|two-component]] response regulator ([[transcription factors of the OmpR family|OmpR family]]) |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of [[phosphate metabolism]] <br/>(''[[phoA]], [[phoB]], [[phoD]], [[resA | resABCDE]], [[tagA]]-[[tagB]], [[tagD | tagDEF]], [[tuaA | tuaA-H]]'') | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of [[phosphate metabolism]] <br/>(''[[phoA]], [[phoB]], [[phoD]], [[resA | resABCDE]], [[tagA]]-[[tagB]], [[tagD | tagDEF]], [[tuaA | tuaA-H]]'') | ||
| Line 41: | Line 41: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | + | <br/><br/> | |
| − | |||
| − | |||
| − | |||
| − | |||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
| Line 85: | Line 81: | ||
* '''Catalyzed reaction/ biological activity:''' | * '''Catalyzed reaction/ biological activity:''' | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' [[transcription factors of the OmpR family|OmpR family]] of two-component response regulators |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
Revision as of 11:08, 13 June 2014
- Description: two-component response regulator (OmpR family), regulation of phosphate metabolism
| Gene name | phoP |
| Synonyms | |
| Essential | no |
| Product | two-component response regulator (OmpR family) |
| Function | regulation of phosphate metabolism (phoA, phoB, phoD, resABCDE, tagA-tagB, tagDEF, tuaA-H) |
| Gene expression levels in SubtiExpress: phoP | |
| Interactions involving this protein in SubtInteract: PhoP | |
| Metabolic function and regulation of this protein in SubtiPathways: phoP | |
| MW, pI | 27 kDa, 5.068 |
| Gene length, protein length | 720 bp, 240 aa |
| Immediate neighbours | phoR, mdh |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 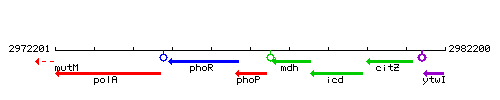
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
phosphate metabolism, transcription factors and their control, regulators of core metabolism, sporulation proteins, general stress proteins (controlled by SigB), membrane proteins, phosphoproteins
This gene is a member of the following regulons
CcpA regulon, PhoP regulon, SigB regulon, SigE regulon
The PhoP regulon
The gene
Basic information
- Locus tag: BSU29110
Phenotypes of a mutant
Database entries
- BsubCyc: BSU29110
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: OmpR family of two-component response regulators
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation by PhoR under conditions of phosphate limitation (stimulates DNA-binding activity)
- Cofactor(s):
- Effectors of protein activity: phosphorylation stimulates DNA-binding activity
- Localization: cell membrane (according to Swiss-Prot)
Database entries
- BsubCyc: BSU29110
- Structure: 1MVO (receiver domain)
- UniProt: P13792
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Marion Hulett, University of Illinois at Chicago, USA Homepage
Your additional remarks
References
Regulation of phoP-phoR expression
Bindiya Kaushal, Salbi Paul, F Marion Hulett
Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC.
J Bacteriol: 2010, 192(12);3103-13
[PubMed:20382764]
[WorldCat.org]
[DOI]
(I p)
Ankita Puri-Taneja, Salbi Paul, Yinghua Chen, F Marion Hulett
CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6).
J Bacteriol: 2006, 188(4);1266-78
[PubMed:16452408]
[WorldCat.org]
[DOI]
(P p)
Salbi Paul, Stephanie Birkey, Wei Liu, F Marion Hulett
Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP.
J Bacteriol: 2004, 186(13);4262-75
[PubMed:15205429]
[WorldCat.org]
[DOI]
(P p)
Zoltán Prágai, Nicholas E E Allenby, Nicola O'Connor, Sarah Dubrac, Georges Rapoport, Tarek Msadek, Colin R Harwood
Transcriptional regulation of the phoPR operon in Bacillus subtilis.
J Bacteriol: 2004, 186(4);1182-90
[PubMed:14762014]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Biochemical analyses
Targets of PhoR
Additional publications: PubMed
Other original publications