Difference between revisions of "KinA"
| Line 61: | Line 61: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU13990&redirect=T BSU13990] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/kinA.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/kinA.html] | ||
| Line 111: | Line 112: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU13990&redirect=T BSU13990] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2VLG 2VLG] (PAS domain) | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2VLG 2VLG] (PAS domain) | ||
Revision as of 13:34, 2 April 2014
- Description: two-component sensor kinase, phosphorylates Spo0F, part of the phosphorelay
| Gene name | kinA |
| Synonyms | spoIIF, spoIIJ, gsiC, scoB, scoD |
| Essential | no |
| Product | two-component sensor kinase |
| Function | initiation of sporulation |
| Gene expression levels in SubtiExpress: kinA | |
| Interactions involving this protein in SubtInteract: KinA | |
| Function and regulation of this protein in SubtiPathways: kinA | |
| MW, pI | 68 kDa, 5.491 |
| Gene length, protein length | 1818 bp, 606 aa |
| Immediate neighbours | pbpH, patA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 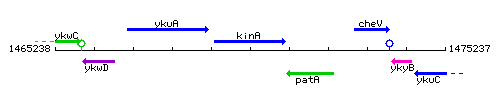
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed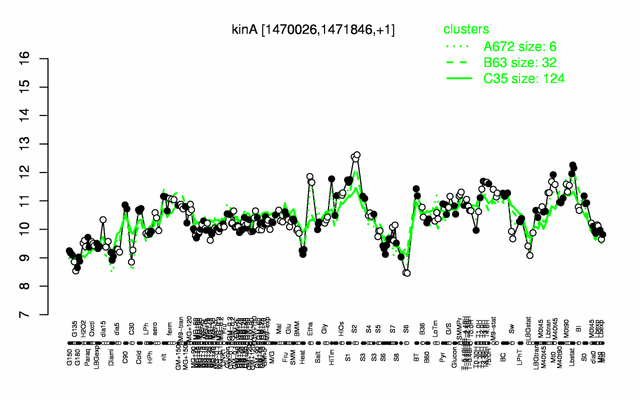
| |
Contents
Categories containing this gene/protein
protein modification, transcription factors and their control, phosphorelay, phosphoproteins
This gene is a member of the following regulons
SigH regulon, Spo0A regulon, stringent response
The gene
Basic information
- Locus tag: BSU13990
Phenotypes of a mutant
Database entries
- BsubCyc: BSU13990
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: autophosphorylation on a His residue
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm
Database entries
- BsubCyc: BSU13990
- Structure: 2VLG (PAS domain)
- UniProt: P16497
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- expressed under conditions that trigger sporulation (Spo0A) PubMed
- induced upon addition of decoyinine (positive stringent response) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Ilana Kolodkin-Gal, Alexander K W Elsholz, Christine Muth, Peter R Girguis, Roberto Kolter, Richard Losick
Respiration control of multicellularity in Bacillus subtilis by a complex of the cytochrome chain with a membrane-embedded histidine kinase.
Genes Dev: 2013, 27(8);887-99
[PubMed:23599347]
[WorldCat.org]
[DOI]
(I p)
Brit Winnen, Eric Anderson, James L Cole, Glenn F King, Susan L Rowland
Role of the PAS sensor domains in the Bacillus subtilis sporulation kinase KinA.
J Bacteriol: 2013, 195(10);2349-58
[PubMed:23504013]
[WorldCat.org]
[DOI]
(I p)
Shigeo Tojo, Kazutake Hirooka, Yasutaro Fujita
Expression of kinA and kinB of Bacillus subtilis, necessary for sporulation initiation, is under positive stringent transcription control.
J Bacteriol: 2013, 195(8);1656-65
[PubMed:23378509]
[WorldCat.org]
[DOI]
(I p)
Sharon Garti-Levi, Ashlee Eswara, Yoav Smith, Masaya Fujita, Sigal Ben-Yehuda
Novel modulators controlling entry into sporulation in Bacillus subtilis.
J Bacteriol: 2013, 195(7);1475-83
[PubMed:23335417]
[WorldCat.org]
[DOI]
(I p)
Jatin Narula, Seram N Devi, Masaya Fujita, Oleg A Igoshin
Ultrasensitivity of the Bacillus subtilis sporulation decision.
Proc Natl Acad Sci U S A: 2012, 109(50);E3513-22
[PubMed:23169620]
[WorldCat.org]
[DOI]
(I p)
Angel E Dago, Alexander Schug, Andrea Procaccini, James A Hoch, Martin Weigt, Hendrik Szurmant
Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis.
Proc Natl Acad Sci U S A: 2012, 109(26);E1733-42
[PubMed:22670053]
[WorldCat.org]
[DOI]
(I p)
Joe H Levine, Michelle E Fontes, Jonathan Dworkin, Michael B Elowitz
Pulsed feedback defers cellular differentiation.
PLoS Biol: 2012, 10(1);e1001252
[PubMed:22303282]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Ashlee Dravis, Seram Nganbiton Devi, Monika Vishnoi, Hoang-Anh Dao, Masaya Fujita
Expression level of a chimeric kinase governs entry into sporulation in Bacillus subtilis.
J Bacteriol: 2011, 193(22);6113-22
[PubMed:21926229]
[WorldCat.org]
[DOI]
(I p)
Anna L McLoon, Ilana Kolodkin-Gal, Shmuel M Rubinstein, Roberto Kolter, Richard Losick
Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis.
J Bacteriol: 2011, 193(3);679-85
[PubMed:21097618]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Daniel Duan, Jeffrey Dinh, Ashlee Dravis, Seram Nganbiton Devi, Masaya Fujita
The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis.
J Bacteriol: 2010, 192(15);3870-82
[PubMed:20511506]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Jeffrey Dinh, Daniel Duan, Oleg A Igoshin, Masaya Fujita
Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells.
Microbiology (Reading): 2010, 156(Pt 8);2294-2304
[PubMed:20413551]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Masaya Fujita
Systematic domain deletion analysis of the major sporulation kinase in Bacillus subtilis.
J Bacteriol: 2010, 192(6);1744-8
[PubMed:20081035]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Tao Guo, Masaya Fujita
In vivo domain-based functional analysis of the major sporulation sensor kinase, KinA, in Bacillus subtilis.
J Bacteriol: 2009, 191(17);5358-68
[PubMed:19561131]
[WorldCat.org]
[DOI]
(I p)
Kazuo Kobayashi, Ritsuko Kuwana, Hiromu Takamatsu
kinA mRNA is missing a stop codon in the undomesticated Bacillus subtilis strain ATCC 6051.
Microbiology (Reading): 2008, 154(Pt 1);54-63
[PubMed:18174125]
[WorldCat.org]
[DOI]
(P p)
Andrew E Whitten, David A Jacques, Boualem Hammouda, Tracey Hanley, Glenn F King, J Mitchell Guss, Jill Trewhella, David B Langley
The structure of the KinA-Sda complex suggests an allosteric mechanism of histidine kinase inhibition.
J Mol Biol: 2007, 368(2);407-20
[PubMed:17350039]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, Richard Losick
Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A.
Genes Dev: 2005, 19(18);2236-44
[PubMed:16166384]
[WorldCat.org]
[DOI]
(P p)
Susan L Rowland, William F Burkholder, Katherine A Cunningham, Mark W Maciejewski, Alan D Grossman, Glenn F King
Structure and mechanism of action of Sda, an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis.
Mol Cell: 2004, 13(5);689-701
[PubMed:15023339]
[WorldCat.org]
[DOI]
(P p)
Sophie J Stephenson, Marta Perego
Interaction surface of the Spo0A response regulator with the Spo0E phosphatase.
Mol Microbiol: 2002, 44(6);1455-67
[PubMed:12067336]
[WorldCat.org]
[DOI]
(P p)
K Stephenson, J A Hoch
PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis.
Proc Natl Acad Sci U S A: 2001, 98(26);15251-6
[PubMed:11734624]
[WorldCat.org]
[DOI]
(P p)
L Wang, C Fabret, K Kanamaru, K Stephenson, V Dartois, M Perego, J A Hoch
Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis.
J Bacteriol: 2001, 183(9);2795-802
[PubMed:11292798]
[WorldCat.org]
[DOI]
(P p)
M Jiang, W Shao, M Perego, J A Hoch
Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis.
Mol Microbiol: 2000, 38(3);535-42
[PubMed:11069677]
[WorldCat.org]
[DOI]
(P p)
C Fabret, V A Feher, J A Hoch
Two-component signal transduction in Bacillus subtilis: how one organism sees its world.
J Bacteriol: 1999, 181(7);1975-83
[PubMed:10094672]
[WorldCat.org]
[DOI]
(P p)
M Fujita, Y Sadaie
Feedback loops involving Spo0A and AbrB in in vitro transcription of the genes involved in the initiation of sporulation in Bacillus subtilis.
J Biochem: 1998, 124(1);98-104
[PubMed:9644251]
[WorldCat.org]
[DOI]
(P p)
C E Grimshaw, S Huang, C G Hanstein, M A Strauch, D Burbulys, L Wang, J A Hoch, J M Whiteley
Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis.
Biochemistry: 1998, 37(5);1365-75
[PubMed:9477965]
[WorldCat.org]
[DOI]
(P p)
L Wang, R Grau, M Perego, J A Hoch
A novel histidine kinase inhibitor regulating development in Bacillus subtilis.
Genes Dev: 1997, 11(19);2569-79
[PubMed:9334321]
[WorldCat.org]
[DOI]
(P p)
Y L Tzeng, J A Hoch
Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis.
J Mol Biol: 1997, 272(2);200-12
[PubMed:9299348]
[WorldCat.org]
[DOI]
(P p)
M Predich, G Nair, I Smith
Bacillus subtilis early sporulation genes kinA, spo0F, and spo0A are transcribed by the RNA polymerase containing sigma H.
J Bacteriol: 1992, 174(9);2771-8
[PubMed:1569009]
[WorldCat.org]
[DOI]
(P p)
D Burbulys, K A Trach, J A Hoch
Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay.
Cell: 1991, 64(3);545-52
[PubMed:1846779]
[WorldCat.org]
[DOI]
(P p)
M Perego, S P Cole, D Burbulys, K Trach, J A Hoch
Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis.
J Bacteriol: 1989, 171(11);6187-96
[PubMed:2509430]
[WorldCat.org]
[DOI]
(P p)