Difference between revisions of "SpoIIE"
| Line 58: | Line 58: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00640&redirect=T BSU00640] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/spoIIE.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/spoIIE.html] | ||
| Line 101: | Line 102: | ||
** localizes to the polar cell division sites where it causes [[FtsZ]] to relocate from mid-cell to form polar Z-rings | ** localizes to the polar cell division sites where it causes [[FtsZ]] to relocate from mid-cell to form polar Z-rings | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00640&redirect=T BSU00640] | ||
* '''Structure:''' | * '''Structure:''' | ||
Revision as of 12:47, 2 April 2014
- Description: protein serine phosphatase, septum-associated PP2C, dephosphorylation of SpoIIAA
| Gene name | spoIIE |
| Synonyms | spoIIH, spoIIK |
| Essential | no |
| Product | protein serine phosphatase, septum-associated PP2C |
| Function | control of SigF activity required for normal formation of the asymmetric septum |
| Gene expression levels in SubtiExpress: spoIIE | |
| Interactions involving this protein in SubtInteract: SpoIIE | |
| Metabolic function and regulation of this protein in SubtiPathways: spoIIE | |
| MW, pI | 91 kDa, 6.137 |
| Gene length, protein length | 2481 bp, 827 aa |
| Immediate neighbours | trnSL-Glu1, yabS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 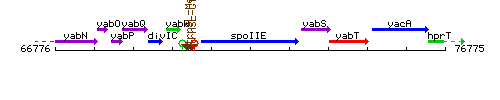
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed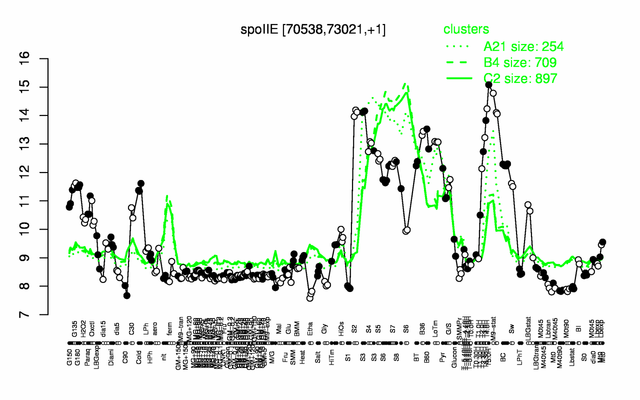
| |
Contents
Categories containing this gene/protein
protein modification, sigma factors and their control, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00640
Phenotypes of a mutant
Database entries
- BsubCyc: BSU00640
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: dephosphorylation of SpoIIAA
- Protein family: PP2C phosphatase
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cell membrane (according to Swiss-Prot)
- localizes to the polar cell division sites where it causes FtsZ to relocate from mid-cell to form polar Z-rings
Database entries
- BsubCyc: BSU00640
- KEGG entry: [3]
- E.C. number: 3.1.3.16
Additional information
Expression and regulation
- Operon: spoIIE (DBTBS)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
Your additional remarks
References
Modeling of SigF activation
Original Publications
Vladimir M Levdikov, Elena V Blagova, Andrea E Rawlings, Katie Jameson, James Tunaley, Darren J Hart, Imrich Barak, Anthony J Wilkinson
Structure of the phosphatase domain of the cell fate determinant SpoIIE from Bacillus subtilis.
J Mol Biol: 2012, 415(2);343-58
[PubMed:22115775]
[WorldCat.org]
[DOI]
(I p)
Andrea E Rawlings, Vladimir M Levdikov, Elena Blagova, Vicki L Colledge, Philippe J Mas, James Tunaley, Ludmila Vavrova, Keith S Wilson, Imrich Barak, Darren J Hart, Anthony J Wilkinson
Expression of soluble, active fragments of the morphogenetic protein SpoIIE from Bacillus subtilis using a library-based construct screen.
Protein Eng Des Sel: 2010, 23(11);817-25
[PubMed:20817757]
[WorldCat.org]
[DOI]
(I p)
Avigdor Eldar, Vasant K Chary, Panagiotis Xenopoulos, Michelle E Fontes, Oliver C Losón, Jonathan Dworkin, Patrick J Piggot, Michael B Elowitz
Partial penetrance facilitates developmental evolution in bacteria.
Nature: 2009, 460(7254);510-4
[PubMed:19578359]
[WorldCat.org]
[DOI]
(I p)
Nathalie Campo, Kathleen A Marquis, David Z Rudner
SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis.
J Biol Chem: 2008, 283(8);4975-82
[PubMed:18077456]
[WorldCat.org]
[DOI]
(P p)
Oleg A Igoshin, Chester W Price, Michael A Savageau
Signalling network with a bistable hysteretic switch controls developmental activation of the sigma transcription factor in Bacillus subtilis.
Mol Microbiol: 2006, 61(1);165-84
[PubMed:16824103]
[WorldCat.org]
[DOI]
(P p)
Shonna M McBride, Aileen Rubio, Lei Wang, William G Haldenwang
Contributions of protein structure and gene position to the compartmentalization of the regulatory proteins sigma(E) and SpoIIE in sporulating Bacillus subtilis.
Mol Microbiol: 2005, 57(2);434-51
[PubMed:15978076]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Sigal Ben-Yehuda, Nicole King, Richard Losick
Genetic dissection of the sporulation protein SpoIIE and its role in asymmetric division in Bacillus subtilis.
J Bacteriol: 2005, 187(10);3511-20
[PubMed:15866939]
[WorldCat.org]
[DOI]
(P p)
Masaya Fujita, José Eduardo González-Pastor, Richard Losick
High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis.
J Bacteriol: 2005, 187(4);1357-68
[PubMed:15687200]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Tae-Jong Kim, Chester W Price, Richard Losick
Insulation of the sigmaF regulatory system in Bacillus subtilis.
J Bacteriol: 2004, 186(13);4390-4
[PubMed:15205443]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Patrick Eichenberger, Richard Losick
A threshold mechanism governing activation of the developmental regulatory protein sigma F in Bacillus subtilis.
J Biol Chem: 2004, 279(15);14860-70
[PubMed:14744853]
[WorldCat.org]
[DOI]
(P p)
Andrea Feucht, Laura Abbotts, Jeffery Errington
The cell differentiation protein SpoIIE contains a regulatory site that controls its phosphatase activity in response to asymmetric septation.
Mol Microbiol: 2002, 45(4);1119-30
[PubMed:12180929]
[WorldCat.org]
[DOI]
(P p)
Sigal Ben-Yehuda, Richard Losick
Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ.
Cell: 2002, 109(2);257-66
[PubMed:12007411]
[WorldCat.org]
[DOI]
(P p)
J Clarkson, I D Campbell, M D Yudkin
NMR studies of the interactions of SpoIIAA with its partner proteins that regulate sporulation in Bacillus subtilis.
J Mol Biol: 2001, 314(3);359-64
[PubMed:11846550]
[WorldCat.org]
[DOI]
(P p)
I Lucet, A Feucht, M D Yudkin, J Errington
Direct interaction between the cell division protein FtsZ and the cell differentiation protein SpoIIE.
EMBO J: 2000, 19(7);1467-75
[PubMed:10747015]
[WorldCat.org]
[DOI]
(P p)
A Feucht, R A Daniel, J Errington
Characterization of a morphological checkpoint coupling cell-specific transcription to septation in Bacillus subtilis.
Mol Microbiol: 1999, 33(5);1015-26
[PubMed:10476035]
[WorldCat.org]
[DOI]
(P p)
N King, O Dreesen, P Stragier, K Pogliano, R Losick
Septation, dephosphorylation, and the activation of sigmaF during sporulation in Bacillus subtilis.
Genes Dev: 1999, 13(9);1156-67
[PubMed:10323866]
[WorldCat.org]
[DOI]
(P p)
I Lucet, R Borriss, M D Yudkin
Purification, kinetic properties, and intracellular concentration of SpoIIE, an integral membrane protein that regulates sporulation in Bacillus subtilis.
J Bacteriol: 1999, 181(10);3242-5
[PubMed:10322028]
[WorldCat.org]
[DOI]
(P p)
N Frandsen, I Barák, C Karmazyn-Campelli, P Stragier
Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis.
Genes Dev: 1999, 13(4);394-9
[PubMed:10049355]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, L J Wu, J Errington
Establishment of prespore-specific gene expression in Bacillus subtilis: localization of SpoIIE phosphatase and initiation of compartment-specific proteolysis.
J Bacteriol: 1998, 180(13);3276-84
[PubMed:9642177]
[WorldCat.org]
[DOI]
(P p)
L J Wu, A Feucht, J Errington
Prespore-specific gene expression in Bacillus subtilis is driven by sequestration of SpoIIE phosphatase to the prespore side of the asymmetric septum.
Genes Dev: 1998, 12(9);1371-80
[PubMed:9573053]
[WorldCat.org]
[DOI]
(P p)
A Khvorova, L Zhang, M L Higgins, P J Piggot
The spoIIE locus is involved in the Spo0A-dependent switch in the location of FtsZ rings in Bacillus subtilis.
J Bacteriol: 1998, 180(5);1256-60
[PubMed:9495766]
[WorldCat.org]
[DOI]
(P p)
P A Levin, R Losick, P Stragier, F Arigoni
Localization of the sporulation protein SpoIIE in Bacillus subtilis is dependent upon the cell division protein FtsZ.
Mol Microbiol: 1997, 25(5);839-46
[PubMed:9364910]
[WorldCat.org]
[DOI]
(P p)
K Pogliano, A E Hofmeister, R Losick
Disappearance of the sigma E transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis.
J Bacteriol: 1997, 179(10);3331-41
[PubMed:9150232]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, T Magnin, J Errington
Compartmentalized distribution of the proteins controlling the prespore-specific transcription factor sigmaF of Bacillus subtilis.
Genes Cells: 1996, 1(10);881-94
[PubMed:9077448]
[WorldCat.org]
[DOI]
(P p)
F Arigoni, L Duncan, S Alper, R Losick, P Stragier
SpoIIE governs the phosphorylation state of a protein regulating transcription factor sigma F during sporulation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1996, 93(8);3238-42
[PubMed:8622920]
[WorldCat.org]
[DOI]
(P p)
A Feucht, T Magnin, M D Yudkin, J Errington
Bifunctional protein required for asymmetric cell division and cell-specific transcription in Bacillus subtilis.
Genes Dev: 1996, 10(7);794-803
[PubMed:8846916]
[WorldCat.org]
[DOI]
(P p)
L Duncan, S Alper, F Arigoni, R Losick, P Stragier
Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division.
Science: 1995, 270(5236);641-4
[PubMed:7570023]
[WorldCat.org]
[DOI]
(P p)
F Arigoni, K Pogliano, C D Webb, P Stragier, R Losick
Localization of protein implicated in establishment of cell type to sites of asymmetric division.
Science: 1995, 270(5236);637-40
[PubMed:7570022]
[WorldCat.org]
[DOI]
(P p)
K York, T J Kenney, S Satola, C P Moran, H Poth, P Youngman
Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE.
J Bacteriol: 1992, 174(8);2648-58
[PubMed:1556084]
[WorldCat.org]
[DOI]
(P p)
P Margolis, A Driks, R Losick
Establishment of cell type by compartmentalized activation of a transcription factor.
Science: 1991, 254(5031);562-5
[PubMed:1948031]
[WorldCat.org]
[DOI]
(P p)