Difference between revisions of "EfeB"
(→Original publications) |
|||
| Line 162: | Line 162: | ||
<pubmed> 24140208 </pubmed> | <pubmed> 24140208 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>16672620,19180538,12354229, 19383693 9353933 9683469 15554971 21479178 18179421 22923395 23180473 23560556 23764491 23820555 </pubmed> | + | <pubmed>16672620,19180538,12354229, 19383693 9353933 9683469 15554971 21479178 18179421 22923395 23180473 23560556 23764491 23820555 24620988 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:38, 14 March 2014
- Description: elemental iron uptake system, heme peroxidase, converts ferrous iron (Fe(II) to ferric iron (FeIII)) for uptake by EfeO-EfeU, peroxide detoxification under microaerobic conditions
| Gene name | efeB |
| Synonyms | ipa-29d, ywbN |
| Essential | no |
| Product | heme peroxidase in elemental iron uptake |
| Function | ferrous iron conversion |
| Gene expression levels in SubtiExpress: efeB | |
| Interactions involving this protein in SubtInteract: EfeB | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 45 kDa, 8.64 |
| Gene length, protein length | 1248 bp, 416 aa |
| Immediate neighbours | ywbO, efeO |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 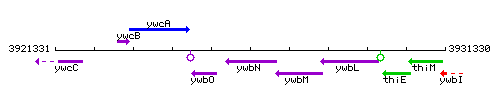
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
acquisition of iron, iron metabolism, cell envelope stress proteins (controlled by SigM, V, W, X, Y), resistance against oxidative and electrophile stress, membrane proteins
This gene is a member of the following regulons
Fur regulon, SigM regulon, SigW regulon, SigX regulon
The gene
Basic information
- Locus tag: BSU38260
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: DyP-type peroxidase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39597
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Jan Maarten van Dijl, Groningen, Netherlands
Your additional remarks
References
Reviews
Original publications