Difference between revisions of "GmuC"
| Line 17: | Line 17: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=GmuC GmuC] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=GmuC GmuC] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=gmuC gmuC]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 48 kDa, 9.803 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 48 kDa, 9.803 | ||
Revision as of 10:14, 7 January 2014
- Description: glucomannan-specific phosphotransferase system, EIIC component of the PTS
| Gene name | gmuC |
| Synonyms | ydhO |
| Essential | no |
| Product | glucomannan-specific phosphotransferase system, EIIC component |
| Function | glucomannan uptake and phosphorylation |
| Gene expression levels in SubtiExpress: gmuC | |
| Interactions involving this protein in SubtInteract: GmuC | |
| Metabolic function and regulation of this protein in SubtiPathways: gmuC | |
| MW, pI | 48 kDa, 9.803 |
| Gene length, protein length | 1326 bp, 442 aa |
| Immediate neighbours | gmuA, gmuD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 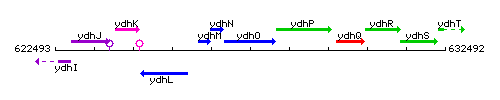
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed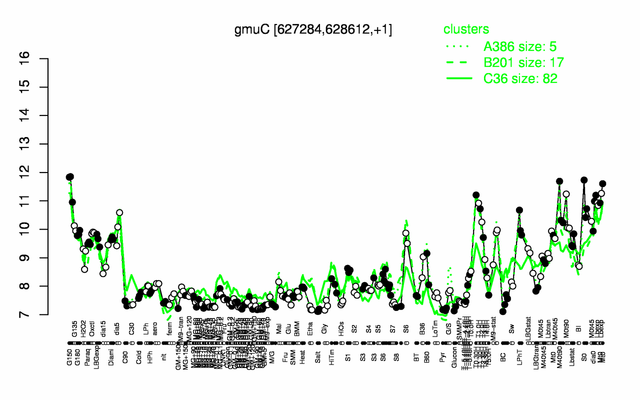
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources
This gene is a member of the following regulons
AbrB regulon, CcpA regulon, GmuR regulon, Efp-dependent proteins
The gene
Basic information
- Locus tag: BSU05830
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 3QNQ (EIIC component of diacetylchitobiose specific PTS from Bacillus cereus; 41% identity, 80% similarity) PubMed
- UniProt: O05507
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
- translation is likely to require Efp due to the presence of several consecutive proline residues PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yu Cao, Xiangshu Jin, Elena J Levin, Hua Huang, Yinong Zong, Matthias Quick, Jun Weng, Yaping Pan, James Love, Marco Punta, Burkhard Rost, Wayne A Hendrickson, Jonathan A Javitch, Kanagalaghatta R Rajashankar, Ming Zhou
Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature: 2011, 473(7345);50-4
[PubMed:21471968]
[WorldCat.org]
[DOI]
(I p)
Onuma Chumsakul, Hiroki Takahashi, Taku Oshima, Takahiro Hishimoto, Shigehiko Kanaya, Naotake Ogasawara, Shu Ishikawa
Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation.
Nucleic Acids Res: 2011, 39(2);414-28
[PubMed:20817675]
[WorldCat.org]
[DOI]
(I p)
Yoshito Sadaie, Hisashi Nakadate, Reiko Fukui, Lii Mien Yee, Kei Asai
Glucomannan utilization operon of Bacillus subtilis.
FEMS Microbiol Lett: 2008, 279(1);103-9
[PubMed:18177310]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)