Difference between revisions of "TatAD"
| Line 84: | Line 84: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 153: | Line 153: | ||
=References= | =References= | ||
== Reviews == | == Reviews == | ||
| − | <pubmed> 22683878 </pubmed> | + | <pubmed> 22683878 24140208 </pubmed> |
== Original publications == | == Original publications == | ||
<pubmed>12867413,15554971,20726548 ,11007775, 19395490 21479178 21479530 22383849 10094677 23180473 23567937 20977272</pubmed> | <pubmed>12867413,15554971,20726548 ,11007775, 19395490 21479178 21479530 22383849 10094677 23180473 23567937 20977272</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 14:07, 6 December 2013
- Description: component of the TatAD-TatCD twin-arginine translocase
| Gene name | tatAD |
| Synonyms | yczB |
| Essential | no |
| Product | component of the twin-arginine translocation pathway |
| Function | TAT protein secretion |
| Gene expression levels in SubtiExpress: tatAD | |
| Interactions involving this protein in SubtInteract: TatAD | |
| Metabolic function and regulation of this protein in SubtiPathways: Protein secretion | |
| MW, pI | 7 kDa, 9.693 |
| Gene length, protein length | 210 bp, 70 aa |
| Immediate neighbours | phoD, tatCD |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 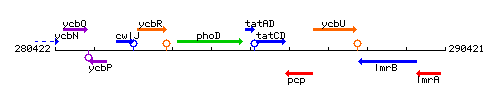
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed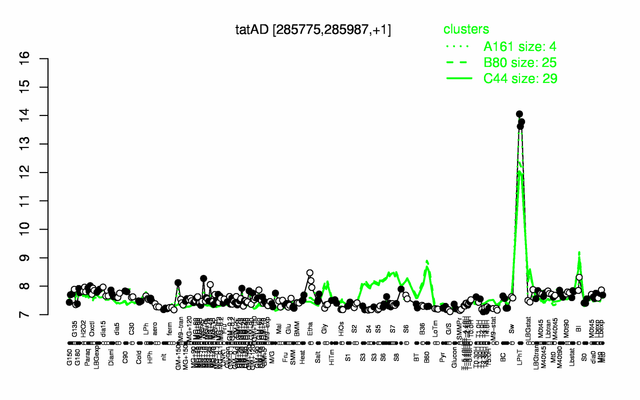
| |
Contents
Categories containing this gene/protein
phosphate metabolism, protein secretion, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02630
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Protein family: tatA/E family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: membrane (according to Swiss-Prot), forms foci at the division sites and cell poles, localization requires interaction with TatCD or TatCY PubMed
Database entries
- UniProt: O31467
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Oscar Kuipers, University of Groningen, The Netherlands Homepage
Your additional remarks
References
Reviews
Vivianne J Goosens, Carmine G Monteferrante, Jan Maarten van Dijl
The Tat system of Gram-positive bacteria.
Biochim Biophys Acta: 2014, 1843(8);1698-706
[PubMed:24140208]
[WorldCat.org]
[DOI]
(P p)
Tracy Palmer, Ben C Berks
The twin-arginine translocation (Tat) protein export pathway.
Nat Rev Microbiol: 2012, 10(7);483-96
[PubMed:22683878]
[WorldCat.org]
[DOI]
(I e)
Original publications