Difference between revisions of "NtdA"
(→Original publications) |
(→The protein) |
||
| Line 82: | Line 82: | ||
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
** pyridoxalphosphate {{PubMed|23586652}} | ** pyridoxalphosphate {{PubMed|23586652}} | ||
| Line 88: | Line 88: | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
| + | ** forms dimers {{PubMed|24097983}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
| Line 93: | Line 94: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' |
| + | ** [http://www.rcsb.org/pdb/explore/explore.do?pdbId=4K2I 4K2I] {{PubMed|24097983}} | ||
* '''UniProt:''' [http://www.uniprot.org/uniprot/O07566 O07566] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O07566 O07566] | ||
Revision as of 19:59, 12 November 2013
- Description: pyridoxal phosphate-dependent 3-oxo-glucose-6-phosphate:glutamate aminotransferase
| Gene name | ntdA |
| Synonyms | yhjL |
| Essential | no |
| Product | pyridoxal phosphate-dependent
3-oxo-glucose-6-phosphate:glutamate aminotransferase |
| Function | synthesis of the antibiotic kanosamine |
| Gene expression levels in SubtiExpress: ntdA | |
| MW, pI | 49 kDa, 5.971 |
| Gene length, protein length | 1323 bp, 441 aa |
| Immediate neighbours | ntdB, ntdR |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 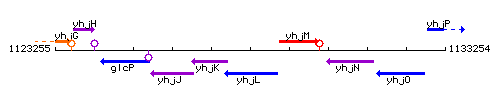
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed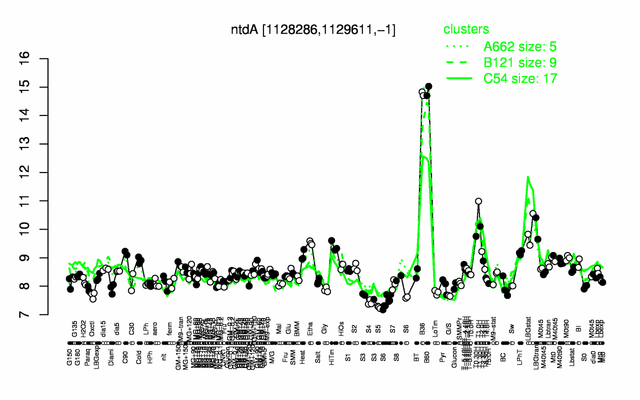
| |
Contents
Categories containing this gene/protein
miscellaneous metabolic pathways, biosynthesis of antibacterial compounds
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10550
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- 3-oxo-d-glucose-6-phosphate + glutamate --> kanosamine-6-phosphate PubMed
- Protein family: degT/dnrJ/eryC1 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Effectors of protein activity:
- Interactions:
- forms dimers PubMed
Database entries
- UniProt: O07566
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Takashi Inaoka, Kozo Ochi
Activation of dormant secondary metabolism neotrehalosadiamine synthesis by an RNA polymerase mutation in Bacillus subtilis.
Biosci Biotechnol Biochem: 2011, 75(4);618-23
[PubMed:21512256]
[WorldCat.org]
[DOI]
(I p)
Original publications