Difference between revisions of "SinI"
(→Original publications) |
|||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || control of [[biofilm formation]] | |style="background:#ABCDEF;" align="center"|'''Function''' || control of [[biofilm formation]] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU24600 sinI] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SinI SinI] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/SinI SinI] | ||
| Line 26: | Line 26: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqhG]]'', ''[[sinR]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yqhG]]'', ''[[sinR]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU24600 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU24600 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU24600 Advanced_DNA] |
|- | |- | ||
|colspan="2" | '''Genetic context''' <br/> [[Image:sinI_context.gif]] | |colspan="2" | '''Genetic context''' <br/> [[Image:sinI_context.gif]] | ||
Revision as of 13:17, 13 May 2013
| Gene name | sinI |
| Synonyms | |
| Essential | no |
| Product | antagonist of SinR |
| Function | control of biofilm formation |
| Gene expression levels in SubtiExpress: sinI | |
| Interactions involving this protein in SubtInteract: SinI | |
| Regulation of this protein in SubtiPathways: Biofilm | |
| MW, pI | 6 kDa, 6.333 |
| Gene length, protein length | 171 bp, 57 aa |
| Immediate neighbours | yqhG, sinR |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 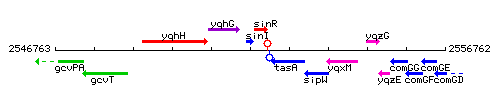
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed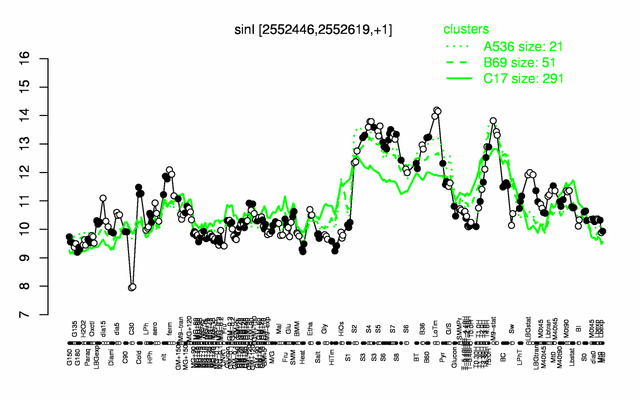
| |
Contents
Categories containing this gene/protein
transcription factors and their control, transition state regulators, biofilm formation
This gene is a member of the following regulons
AbrB regulon, ScoC regulon, Spo0A regulon
The gene
Basic information
- Locus tag: BSU24600
Phenotypes of a mutant
- altered cell death pattern in colonies PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): SlrA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P23308
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- FLAG-tag construct (C-term): GP935 (kan), available in Stülke lab
- Antibody:
Labs working on this gene/ protein
Your additional remarks
References
Reviews
Patrick Piggot
Epigenetic switching: bacteria hedge bets about staying or moving.
Curr Biol: 2010, 20(11);R480-2
[PubMed:20541494]
[WorldCat.org]
[DOI]
(I p)
Modelling of the SinI/SinR switch
Original publications
Joseph A Newman, Cecilia Rodrigues, Richard J Lewis
Molecular basis of the activity of SinR protein, the master regulator of biofilm formation in Bacillus subtilis.
J Biol Chem: 2013, 288(15);10766-78
[PubMed:23430750]
[WorldCat.org]
[DOI]
(I p)
Munehiro Asally, Mark Kittisopikul, Pau Rué, Yingjie Du, Zhenxing Hu, Tolga Çağatay, Andra B Robinson, Hongbing Lu, Jordi Garcia-Ojalvo, Gürol M Süel
Localized cell death focuses mechanical forces during 3D patterning in a biofilm.
Proc Natl Acad Sci U S A: 2012, 109(46);18891-6
[PubMed:23012477]
[WorldCat.org]
[DOI]
(I p)
Martin Lehnik-Habrink, Marc Schaffer, Ulrike Mäder, Christine Diethmaier, Christina Herzberg, Jörg Stülke
RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y.
Mol Microbiol: 2011, 81(6);1459-73
[PubMed:21815947]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Thomas Norman, Roberto Kolter, Richard Losick
Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis.
EMBO J: 2011, 30(7);1402-13
[PubMed:21326214]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Roberto Kolter, Richard Losick
Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability.
Mol Microbiol: 2010, 78(1);218-29
[PubMed:20923420]
[WorldCat.org]
[DOI]
(I p)
Yunrong Chai, Frances Chu, Roberto Kolter, Richard Losick
Bistability and biofilm formation in Bacillus subtilis.
Mol Microbiol: 2008, 67(2);254-63
[PubMed:18047568]
[WorldCat.org]
[DOI]
(P p)
Daniel B Kearns, Frances Chu, Steven S Branda, Roberto Kolter, Richard Losick
A master regulator for biofilm formation by Bacillus subtilis.
Mol Microbiol: 2005, 55(3);739-49
[PubMed:15661000]
[WorldCat.org]
[DOI]
(P p)
Alejandro Sánchez, Jorge Olmos
Bacillus subtilis transcriptional regulators interaction.
Biotechnol Lett: 2004, 26(5);403-7
[PubMed:15104138]
[WorldCat.org]
[DOI]
(P p)
Sasha H Shafikhani, Ines Mandic-Mulec, Mark A Strauch, Issar Smith, Terrance Leighton
Postexponential regulation of sin operon expression in Bacillus subtilis.
J Bacteriol: 2002, 184(2);564-71
[PubMed:11751836]
[WorldCat.org]
[DOI]
(P p)
D J Scott, S Leejeerajumnean, J A Brannigan, R J Lewis, A J Wilkinson, J G Hoggett
Quaternary re-arrangement analysed by spectral enhancement: the interaction of a sporulation repressor with its antagonist.
J Mol Biol: 1999, 293(5);997-1004
[PubMed:10547280]
[WorldCat.org]
[DOI]
(P p)
R J Lewis, J A Brannigan, W A Offen, I Smith, A J Wilkinson
An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex.
J Mol Biol: 1998, 283(5);907-12
[PubMed:9799632]
[WorldCat.org]
[DOI]
(P p)
M A Strauch
In vitro binding affinity of the Bacillus subtilis AbrB protein to six different DNA target regions.
J Bacteriol: 1995, 177(15);4532-6
[PubMed:7635837]
[WorldCat.org]
[DOI]
(P p)
U Bai, I Mandic-Mulec, I Smith
SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction.
Genes Dev: 1993, 7(1);139-48
[PubMed:8422983]
[WorldCat.org]
[DOI]
(P p)
P T Kallio, J E Fagelson, J A Hoch, M A Strauch
The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein.
J Biol Chem: 1991, 266(20);13411-7
[PubMed:1906467]
[WorldCat.org]
(P p)
N K Gaur, K Cabane, I Smith
Structure and expression of the Bacillus subtilis sin operon.
J Bacteriol: 1988, 170(3);1046-53
[PubMed:3125149]
[WorldCat.org]
[DOI]
(P p)